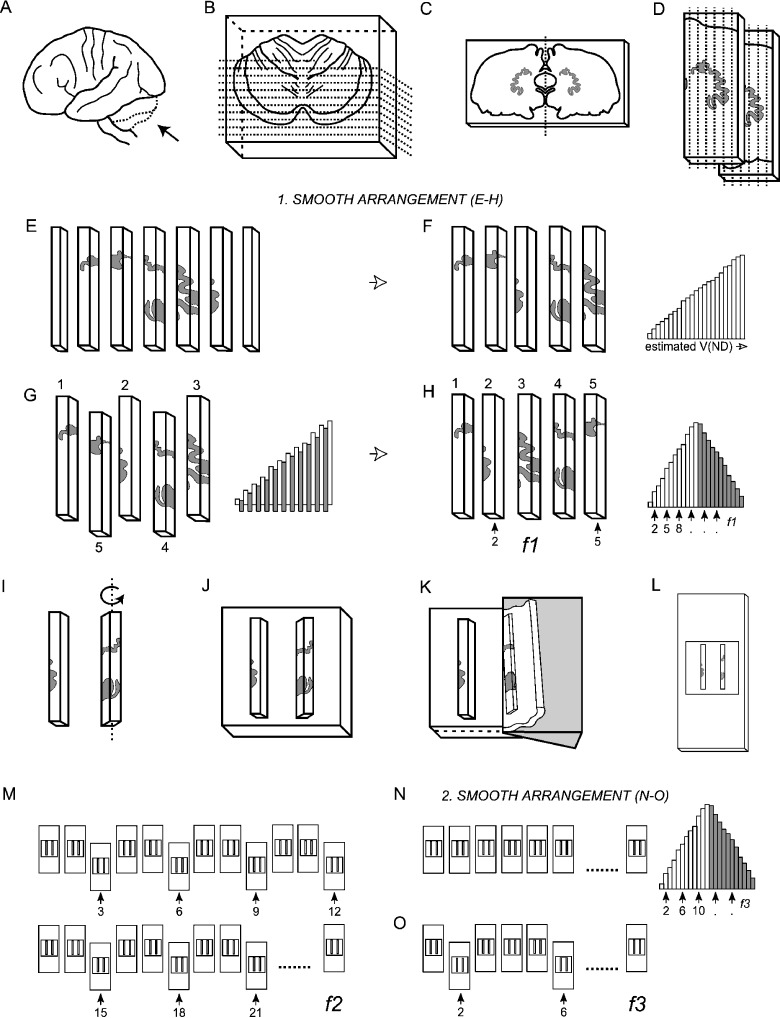
Fig. 2.
The smooth fractionator sampling design. (A) The cerebellum was dissected from the brainstem at the level of the 3rd cranial nerve (B) embedded in agar and sliced coronally into 4-mm-thick, and later 2-mm-thick slices with a random starting position within the slab thickness. (C) The dentate nucleus was identified on 10–12 of the 2-mm-thick slices. (D,E) The left or right dentate nucleus was chosen systematically randomly, dissected and cut into bars. (F) The bars were sorted in an ascending manner according to the amount of DN they contained. (G) From the sorted sequence every second bar was pushed out and rearranged into a smooth arrangement. (H) Depending on sample size a predetermined fraction (f1) was now sampled systematically randomly from the smooth arrangement using a random number table, providing 12–20 bars for further processing. (I) The bars were rotated around their longitudinal axis (see text), (J) embedded in agar, (K) sectioned exhaustively on a vibratome into 100-µm-thick sections, and (L) mounted on microscope slides. (M) From the complete series of sections a predetermined fraction (f2), in this case every 3rd section, was sampled. (N) These sections were again arranged into a smooth fractionator design (O) before a final fraction (f3) was subsampled for cell counting.