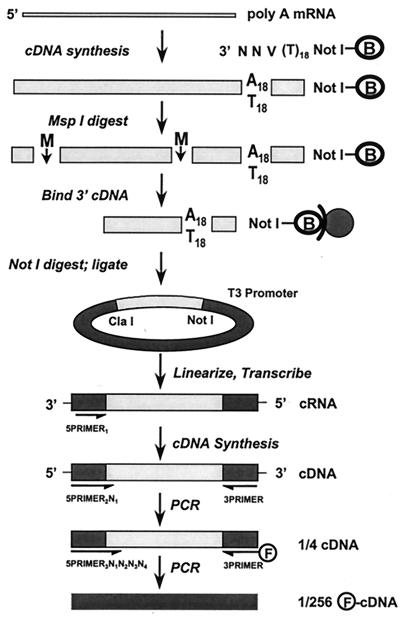
Figure 1.
Schematic view of TOGA. Poly(A)-selected RNA serves as template for double-strand cDNA synthesis by using a pool of NotI-containing biotinylated (B) primers degenerate in their 3′ ultimate three positions that phase the 3′ end of the cDNA at the poly(A) tail. After cleavage with MspI (M), the 3′ biotinylated fragment is captured on streptavidin magnetic beads and released from the beads by digestion with NotI, and the 3′ MspI–NotI fragments are cloned into an RNA expression vector in an orientation antisense to its T3 promoter. After cleavage with MspI to linearize insert-containing plasmids and inactivate insertless plasmids, antisense transcripts are produced with T3 RNA polymerase. These serve, after removal of DNA template, as substrates for reverse transcriptase by using a primer that anneals to vector sequences. A PCR step with a primer that extends across the nonreconstituted MspI/ClaI site by one of the four possible nucleotides and a universal 3′ primer subdivides the cDNA species into four pools. A subsequent PCR in the presence of a fluorescent 3′ primer and each (in separate reactions) of the 256 possible 5′ primers that extends 4 nt into the inserts subdivides the input species into 256 subpools for electrophoretic resolution.