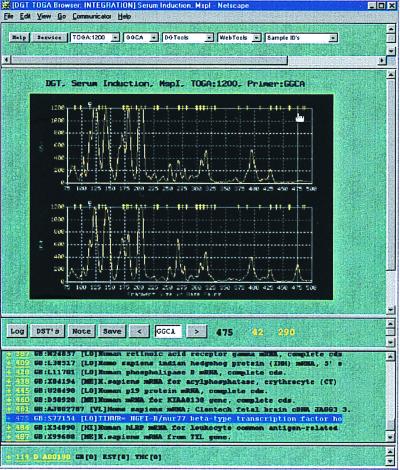
Figure 2.
TOGA data are viewed through a Netscape-based browser. The normalized TOGA profiles appear below five pull-down menus, from left to right: (i) Display Type lists four selections for the y-axis amplitude scale, with ranges from 0 to 600, 1,200, 2,000, or 8,000 normalized fluorescence units (the 1,200 scale is that selected); (ii) Primer lists the 256 permutations of the N1N2N3N4 nucleotides (GGCA is that selected; this selection is also indicated below the display flanked by forward–backward selector buttons for navigating through the alphabetical list and again above the trace display in a title region that also specifies collaborator, experiment name, enzyme identity, and the selected y-axis scale); (iii) DGTools contains selections for searching public and private sequence databases by using blast, gap-blast, or the Keyword search algorithm, for retrieval using the GCG fetch program, for comparing a selected group of sequences using pileup, and for translation of a nucleotide sequence into the putative proteins encoded by its different reading frames (all of these operations are carried out behind a security firewall); (iv) Web Tools gives access to programs and databases on the Internet that require navigation beyond the firewall, including PubMed for medline literature searches and the National Center for Biotechnology Information for searching gene databases; (v) Sample ID's lists the displayed sample identities (here, human osteosarcoma cells deprived of or replenished with serum). A guideline has been placed by mouse on a peak. The length (475 nt) and fragment peak amplitudes (42 and 290 fluorescence units) measurements then automatically appear below the panels. Selection of the save button records the selected peak and its corresponding length and amplitude measurements to a user log file, creating hyperlinks to the original data. Access to that log file is via the button “log.” Below the trace display are the virtual DSTs derived by processing human entries in public databases, arranged by length, and including accession number and entry name and hyperlinked to the GenBank RSF. These virtual DSTs are also mapped above the TOGA traces as yellow hash marks. The 475-nt differentially represented peak selected by the guideline falls under such a hash mark. Consultation of the virtual DST list suggests a candidate identity with TINUR, an NGFI-B/nur77 homologue, which was validated by extended primer PCR and direct nucleotide sequence analysis of the excised fragment.