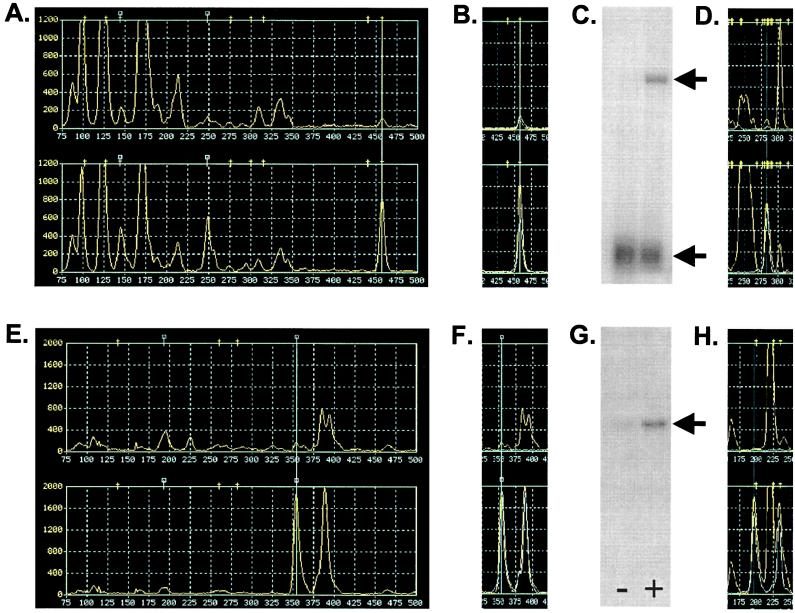
Figure 4.
TOGA candidate verification, fragment capture, and validation. Pool profiles generated from serum-depleted (Upper) and serum-replenished (Lower) cells by N1N2N3N4 primers CCCG (A) and ATCG (E) in the MspI vector; amplitude scales are 1,200 and 2,000, respectively. In A, B, D–F, and H, the guideline is placed over a peak of greater amplitude in the trace from the serum-replenished sample. In A, these peaks comigrate with hash marks corresponding to the virtual DST for NF-κB; in E, there was no comigrating DST (square hash marks represent nonpublic DSTs). In B and F, the region of the traces containing the highlighted peaks is shown overlaid with the products generated by the 14-nt extended primers specific for NF-κB and OS51, respectively, synthesized based on the sequences in the corresponding RSFs (accession nos. M58603 and AF178682). The OS51 extension primer was GATCGAATCCGATCGCTCCTGGTTCCTCGGA. Northern blots for NF-κB (C) and OS51 (G) were prepared from RNA samples from osteosarcoma cells depleted of (−) or replenished with (+) serum. The lower band in C represents the mRNA for ribosomal protein S20, which was used as a normalization standard. Arrows indicate the relevant blot bands. In D and H, the relevant local regions of the corresponding Sau3AI vector panels for each RNA is shown. In E, F, and H, note the alternative poly(A) variant that migrates as 40 nt longer than the OS51 DST.