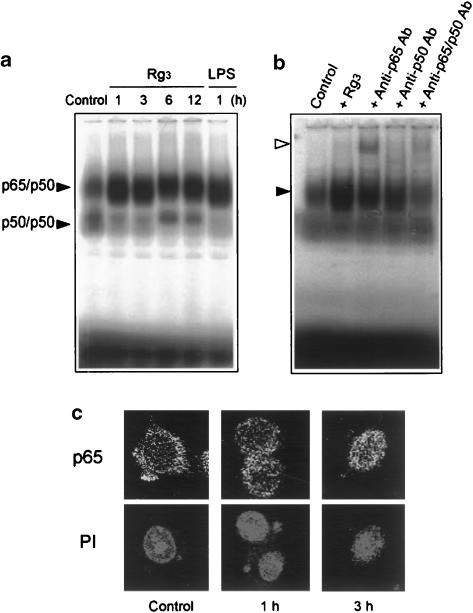
Figure 6.
NF-κB activation by Rg3. (a) Gel shift analysis of NF-κB DNA binding. Gel shift analysis was performed with the nuclear extracts prepared from the Raw264.7 cells incubated with or without Rg3 (10 μg ml−1) for 1–12 h. Each lane contained 2 μg of nuclear extracts. LPS (1 μg ml−1, 1 h) was used for comparative purpose. (b) The specificity of NF-κB DNA binding was confirmed by a decrease in band intensity or supershift with anti-p65 or anti-p50 antibodies. The arrowheads show p65/p50 NF-κB complex (closed) and supershifted NF-κB (open). (c) The nuclear translocation of p65. Immunocytochemistry was performed with the Raw264.7 cells treated with Rg3 (10 μg ml−1) for 1–3 h. p65 migrated toward the nucleus or translocated into the nucleus at 1–3 h. The same fields were counterstained with propidium iodide (PI) to locate the nuclei. LPS, lipopolysaccharide.