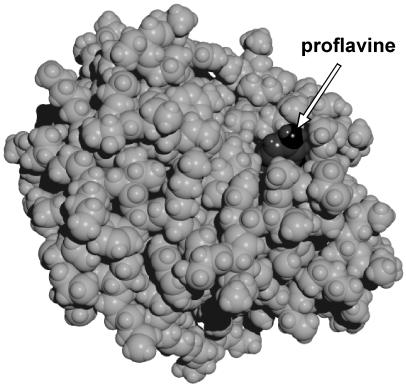
Figure 3.
The proflavine–chymotrypsin complex. The arrow points to the edge of proflavine molecule. The structure was obtained by 10-ps molecular dynamics simulations. Calculations were performed within hyper chem 5.0 (Hypercube, Gainesville, FL) package using the amber (64) force field with 4,000 water molecules in the periodic box of 56 × 50 × 56 Å with the time step of 1 fs. Partial charges of proflavine were obtained from CNDO/2 semiempirical quantum chemical calculations. Other force field parameters were taken from amber parameters corresponding to similar atoms and chemical bonds of nucleotide bases. X-ray coordinates of chymotrypsin atoms with added hydrogen atoms and equilibrated for 20-ps molecular dynamics run without the dye were taken as the initial configuration of the protein.