Abstract
Cyclosporin A and tacrolimus are clinically important immunosuppressive drugs directly targeting the transcription factor nuclear factor of activated T cells (NFAT). Through inhibition of calcineurin phosphatase activity they block the dephosphorylation and thus activation of NFAT. Cyclosporin A and tacrolimus also inhibit other calcineurin-dependent transcription factors including the ubiquitously expressed cAMP response element-binding protein (CREB). Membrane depolarization by phosphorylating CREB on Ser119 leads to the recruitment of its coactivator CREB-binding protein (CBP) that stimulates initiation of transcription.
It was unknown at what step in CREB-mediated transcription cyclosporin A and tacrolimus interfere.
In transient transfection experiments, using GAL4-CREB fusion proteins and a pancreatic islet β-cell line, cyclosporin A inhibited depolarization-induced activation of CREB proteins which carried various deletions or mutations throughout their sequence providing no evidence for the existence of a distinct CREB domain conferring cyclosporin A sensitivity. In a mammalian two-hybrid assay, cyclosporin A did not inhibit Ser119-dependent interaction of CREB with its coactivator CBP.
Using GAL4-CBP fusion proteins, cyclosporin A inhibited depolarization-induced CBP activity, with cyclosporin A-sensitive domains mapped to both the N- (aa 1–451) and C-terminal (aa 2040–2305) ends of CBP. The depolarization-induced transcriptional activity of the CBP C-terminus was enhanced by overexpression of calcineurin and was inhibited by cyclosporin A and tacrolimus in a concentration-dependent manner with IC50 values (10 and 1 nM, respectively) consistent with their known IC50 values for inhibition of calcineurin.
These data suggest that, in contrast to NFAT, cyclosporin A and tacrolimus inhibit CREB transcriptional activity at the coactivator level.
Keywords: Cyclosporin A, cAMP response element-binding protein (CREB), CREB-binding protein (CBP), calcineurin, pancreatic β cell
Introduction
Cyclosporin A and tacrolimus (FK506) are powerful immunosuppressive drugs widely used to prevent organ rejection after transplantation. In addition, the treatment of autoimmune diseases represents a new field for their application. Cyclosporin A and tacrolimus bind to their respective intracellular receptors, the immunophilins, and it is the drug/immunophilin complex that binds to and inhibits the calcium/calmodulin-dependent serine/threonine phosphatase calcineurin (Ho et al., 1996). Since both structurally distinct drugs share the desired immunosuppressive and many undesired effects, it is generally assumed that inhibition of calcineurin is the underlying mechanism (Ho et al., 1996). Through inhibition of calcineurin, cyclosporin A and tacrolimus directly target the transcription factor nuclear factor of activated T cells (NFAT). They prevent the dephosphorylation, nuclear translocation, and thus activation of NFAT, thereby blocking NFAT-dependent gene transcription including IL-2 gene transcription (Ho et al., 1996).
Besides NFAT, cyclosporin A and tacrolimus also inhibit the calcineurin-dependent activity of other transcription factors including the cAMP response element-binding protein (CREB) (Schwaninger et al., 1993a; 1995; Krüger et al., 1997; Oetjen et al., 2003b). CREB is a ubiquitously expressed transcription factor and was shown to play a pivotal role in many different physiological and developmental functions like learning and memory (Mayr & Montminy, 2001; Gau et al., 2002), glucose homeostasis (Herzig et al., 2001; Oetjen et al., 2003b), in growth-factor-dependent cell survival (Mayr & Montminy, 2001), and in neurodegeneration (Mantamadiotis et al., 2002). CREB−/− mice dying at birth from respiratory distress show impaired fetal T-cell development primarily in the α/β lineage and a reduced number of developing T cells in the thymus (Rudolph et al., 1998). In addition, transgenic mice carrying a dominant-negative CREB mutant were defective in T-cell proliferation and activation (Barton et al., 1996). Taken together, these findings strongly suggest that inhibition of CREB transcriptional activity through blocking of calcineurin activity by cyclosporin A and tacrolimus might contribute to the immunosuppressive effect (Barton et al., 1996; Krüger et al., 1997) as well as to untoward effects like impaired glucose tolerance (Oetjen et al., 2003a, 2003b) shared by both drugs.
CREB is a member of the basic region leucine zipper family of transcription factors and binds, typically as a homodimer, to DNA sequences with the consensus core octamer motif TGACGTCA (cAMP response element, CRE), which is present in many genes (Habener et al., 1995; Mayr & Montminy, 2001). The transcriptional activity of CREB is markedly induced by various signalling pathways such as cAMP-, calcium-, and ERK/p38 MAPK-activated signalling pathways through phosphorylation of CREB on Ser119 (in CREB-327, corresponding to Ser133 in CREB-341) (Habener et al., 1995; Mayr & Montminy, 2001). This phosphorylation allows CREB to interact with its coactivator CREB-binding protein (CBP) (Chrivia et al., 1993; Kwok et al., 1994). CBP then mediates CREB transcriptional activity by interacting with components of the general transcription machinery, by chromatin remodelling through its intrinsic histone acetyltransferase activity and by recruiting additional cofactors like pCAF and pCIP (Kwok et al., 1994; Nakajima et al., 1997; Korzus et al., 1998; Kurokawa et al., 1998; Martinez-Balbas et al., 1998). In addition to the phosphorylation on Ser119, CREB transcriptional activity is regulated by multiple signalling pathways acting at different steps of CREB-mediated transcriptional activation: at the level of CREB itself (Sun et al., 1994; Parker et al., 1998; Wu & McMurray, 2001), at the level of CREB–CBP interaction (Sun & Maurer, 1995; Parker et al., 1996; 1998; Mayr et al., 2001; Wu & McMurray, 2001), and at the level of CBP function (Chawla et al., 1998; Gau et al., 2002). It has been shown that cyclosporin A and tacrolimus inhibit CRE-directed transcription after stimulation by membrane depolarization-induced calcium influx or by an elevation of intracellular cAMP levels in various cell lines without decreasing the phosphorylation of CREB on Ser119 (Schwaninger et al., 1993a, 1993b; 1995; Krüger et al., 1997). However, it remained unclear at which step of CREB-mediated transactivation cyclosporin A and tacrolimus interfere.
Using the electrically excitable pancreatic islet β-cell line HIT, the present study shows that cyclosporin A inhibits membrane depolarization-induced CREB activity also after introduction of extensive deletions or mutations into the CREB transactivation domain, providing no evidence for the existence of a distinct cyclosporin A-sensitive CREB domain. Furthermore, in a mammalian two-hybrid assay, cyclosporin A did not disturb the interaction between CREB and CBP. However, cyclosporin A and tacrolimus inhibited membrane depolarization-induced CBP activity, most likely through inhibition of calcineurin. These findings suggest that cyclosporin A and tacrolimus, in contrast to their targeting of NFAT itself, inhibit CREB transcriptional activity at the coactivator level.
Methods
Molecular biology
The reporter gene 5xGal4E1BLuc has been described before (Krüger et al., 1997). pGAL4-CREB-TAD, pGAL4-CREB(1–150), pGAL4-CREB(1–187), pGAL4-CREB(1–204), pGAL4-CREB(92–261), pGAL4-CREB(146–261), pGAL4-CREB(206–261), pGAL4-CREB(Δ92–108), pGAL4-CREB(Δ121–131), pGAL4-CREB(Δ138–148) were a kind gift from C. Lee and J.F. Habener (Boston, MA, U.S.A.); pGAL4-CREB wild-type, containing the transactivation domain of CREB-341 from amino acid(s) (aa) 4 to 283, was like pGAL4-CREB-S133A, and pGAL4-CREB-S142A a kind gift from R.A. Maurer (Portland, OR, U.S.A.) (Sun et al., 1994); pHKnVP16 containing the transactivation domain of VP16 was a kind gift from A. Bannister (Cambridge, U.K.) (Bannister & Kouzarides, 1995); pGAL4-CBP, pGAL4-CBP(442–661) were a kind gift from R.H. Goodman (Portland, OR, U.S.A.) (Kwok et al., 1994); pGAL4-CBP(N), pGAL4-CBP(M), and pGAL4-CBP(C) were a kind gift from C. Glass (San Diego, CA, U.S.A.) (Kurokawa et al., 1998). The expression vector for both calcineurin subunits A and B were a kind gift from G.R. Crabtree (Stanford, CA, U.S.A.) (Clipstone & Crabtree, 1992). The plasmids pHKnCREB/VP16 and pHKnCREB(S119A)/VP16 were generated by PCR using 5′-CCCAAGCTTATGACCATGGAATCTGG-3′ and 5′-CCCAAGCTTAGGAAGTGCTGGGG-3′ as 5′ and 3′ primers, respectively. GAL4-CREB-327 wild-type and GAL4-CREB-327 PKA mut, respectively (kind gift of O.M. Andrisani, West Lafayette, IN, U.S.A.) (Fiol et al., 1994) served as template DNA. The PCR fragments were cloned into the HindIII site of pHKnVP16. The plasmids pGAL4-CBP(1880–2441), pGAL4-CBP(2040–2441), pGAL4-CBP(2171–2441), and pGAL4-CBP(2306–2441) were generated by PCR using 5′-TAGGATCCGTCAGCAGAGTTTGCCT-3′, 5′-TAGGATCCGTGTAATGTCCATGCAG-3′, 5′-TAGGATCCGTGCTGTGAACATCATG-3′, 5′-TAGGATCCGTTCACCAGGCCAGCC-3′, respectively, as 5′ primers and 5′-ACGCGGAGCTCCTACAAACCCTCCAC-3′ as 3′ primer. The plasmid pGAL4-CBP(2040–2305) was generated by PCR using 5′-TAGGATCCGTGTAATGTCCATGCAG-3′ as 5′ primer and 5′-ACGCGGAGCTCCTACCCAATTTGTTGCTTG-3′ as 3′ primer. The resulting PCR fragments were cloned into the BamHI and SacI sites of pSG424, an expression vector coding for the DNA-binding domain of GAL4 from aa 1 to 147 (Sadowski & Ptashne, 1989). For the generation of the internal deletions within the CBP C-terminus, a fragment containing the aa from 1678 to 2039 using 5′-TAGGATCCGTGAATTCTCTTCC-3′ as 5′ primer and 5′-CCAAGCTTTACAGGCCTGGGCATG-3′ as 3′primer and a fragment containing the aa from 2171 to 2441 using 5′-CCCAAGCTTGCTCTGAACATCATGAAC-3′ as 5′ primer and 5′-ACGCGGAGCTCCTACAAACCCTCCAC-3′ as 3′ primer were generated by PCR. For the extended internal deletion, an additional fragment containing the aa from 2306 to 2441 was generated by PCR using 5′-CCCAAGCTTTCACCAGGCCAGCCG-3′ as new 5′ primer. Both resulting PCR fragments were ligated in one step into the BamHI and SacI sites of pSG424. The identity and orientation of all constructs was confirmed by sequencing, using the enzymatic method.
Cell culture and transfection
The pancreatic islet β-cell line HIT (Schwaninger et al., 1993b) was grown in RPMI 1640 supplemented with 10% fetal calf serum, 5% horse serum, 100 U ml−1 penicillin, and 100 μg ml−1 streptomycin. HIT cells were transiently transfected by the DEAE-dextran method (Schwaninger et al., 1993b) with 2 μg of indicator plasmid per 6-cm dish and 2 μg of expression vector per 6-cm dish, unless stated otherwise. Rous sarcoma virus-chloramphenicol acetyltransferase (0.5 μg per 6-cm dish) or cytomegalovirus green fluorescent protein expression vectors (1 μg per 6-cm dish) were added as second reporters to control for transfection efficiency. Cotransfections were carried out with a constant DNA concentration, which was maintained by adding the empty vector. Cells were stimulated with high KCl (final concentration 45 mM) or cAMP (forskolin 10 μM) 6 h before harvest, cyclosporin A or tacrolimus were added 1 h before stimulation. Cell extracts were prepared 48 h after transfection. The chloramphenicol acetyltransferase assay and the luciferase assay were performed as previously described (Schwaninger et al., 1993b). Fluorescence of the green fluorescent protein was measured in a Packard FluoroCount with excitation wavelength at 485 nm and emission wavelength at 530 nm.
Chemicals
Luciferin was purchased from P.J.K. Industrievertretungen (Kleinblittersdorf, Germany), forskolin was obtained from Sigma (Taufkirchen, Germany). Cyclosporin A was provided by Novartis Pharma AG (Basel, Switzerland), tacrolimus (FK506) by Fujisawa (Osaka, Japan). A stock solution of cyclosporin A (10 mg ml−1) was prepared in ethanol with 20% Tween 80 and further diluted in RPMI. Forskolin was solved in dimethyl sulfoxide, tacrolimus in ethanol. Controls received the solvent only.
Statistical assessment
All results are reported as means±standard error of the mean (s.e.m.) for each series of experiments. When indicated, data were analysed by one-factor analysis of variance (ANOVA) and Student's t-test (Statistica, StatSoft(Europe) GmbH, Hamburg, Germany).
Results
Effect of deletions or a mutation within the CREB transactivation domain on the inhibition by cyclosporin A of membrane depolarization-induced CREB transcriptional activity
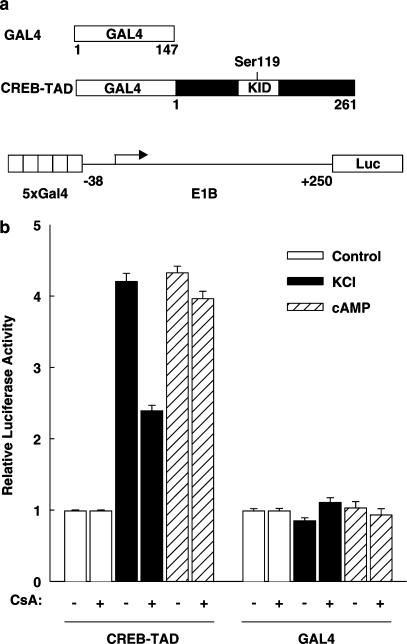
Previous studies have shown that cyclosporin A and tacrolimus inhibit the membrane depolarization-induced transcriptional activity of the CRE and the GAL4-CREB transcriptional activity induced by cAMP plus membrane depolarization (Schwaninger et al., 1993a; 1995; Oetjen et al., 2003b). To investigate the effect of cyclosporin A on the transcriptional activity of the CREB transactivation domain induced by membrane depolarization alone, a luciferase reporter gene under the control of five GAL4-binding sites and the minimal E1B promoter (plasmid 5xGal4E1BLuc) was transiently transfected together with an expression vector for a fusion protein containing the DNA-binding domain of the yeast transcription factor GAL4 (aa 1–147) and the transactivation domain of CREB-327 (aa 1–261) (Figure 1a). The experiments were performed in the electrically excitable pancreatic islet β-cell line HIT, a cell line in which the effect of cyclosporin A and tacrolimus on CRE/CREB-dependent transcription is well characterized (Schwaninger et al., 1993a; 1995; Siemann et al., 1999; Oetjen et al., 2003b). Furthermore, the inhibition of CREB activity by cyclosporin A and tacrolimus in β cells is associated with inhibition of insulin gene transcription that contributes to the undesired diabetogenic effect of these immunosuppressive drugs (Lawrence et al., 2001; Oetjen et al., 2003a, 2003b). As shown in Figure 1b, high KCl-induced membrane depolarization and calcium influx stimulated CREB transactivation four-fold. The adenylate cyclase activator forskolin enhanced transcription to the same extent. Cyclosporin A decreased membrane depolarization-induced transcription mediated by the CREB transactivation domain by 57% (Figure 1b). Transcription stimulated by forskolin was not reduced by cyclosporin A (Figure 1b). Transcription directed by the DNA-binding domain of GAL4 alone was neither enhanced by membrane depolarization or cAMP nor inhibited by cyclosporin A (Figure 1b). These data show that the CREB transactivation domain fused to the DNA-binding domain of GAL4 confers membrane depolarization and cyclosporin A sensitivity. When compared to studies on the transcriptional activity of the CRE (Schwaninger et al., 1993a; 1995) or the GAL4-CREB-full length protein (data not shown), the inhibition by cyclosporin A of the transcriptional activity of the GAL4-CREB transactivation domain fusion protein (Figure 1b) after membrane depolarization or cAMP was less complete or absent, respectively, suggesting that in addition to the isolated CREB transactivation domain other mechanisms, probably depending on the CREB basic region leucine zipper domain, are also involved in conferring cyclosporin A sensitivity. Consistent with this, a newly described protein, the transducer of regulated CREB activity, TORC, was shown to interact with the basic region leucine zipper domain of CREB (Screaton et al., 2004). Besides CREB, the transcriptional activities of the transactivation domains of the highly homologous CRE-binding proteins ATF-1 and CREMτ were also enhanced by membrane depolarization and inhibited by cyclosporin A (data not shown). Thus, in the pancreatic islet β-cell line HIT, the transactivation domains of CREB and the highly homologous ATF-1 and CREMτ proteins confer responsiveness to cyclosporin A after membrane depolarization.
Figure 1.
Cyclosporin A inhibits the transcriptional activity of the CREB transactivation domain after membrane depolarization. (a) Scheme of the GAL4 expression vectors and the luciferase reporter gene used. GAL4 represents the DNA-binding domain of the yeast transcription factor GAL4 (aa 1–147). CREB-TAD, transactivation domain of CREB-327 (aa 1–261) fused to GAL4. KID represents the kinase-inducible domain of CREB. The position of Ser119 is marked. Luc, luciferase. (b) Effect of cyclosporin A. The luciferase reporter gene (5xGal4E1BLuc) and the expression vectors for GAL4 (GAL4) or GAL4 fused to the CREB transactivation domain (CREB-TAD) were transiently cotransfected into HIT cells. Cells were stimulated with high potassium-induced membrane depolarization (KCl, final concentration 45 mM) or 10 μM forskolin (cAMP) 6 h before harvest. Cyclosporin A 5 μM (CsA) was added as indicated 7 h before harvest. Luciferase activity is expressed relative to the mean value in each experiment of the activity measured in the respective control. Values are means±s.e.m. (n=12).
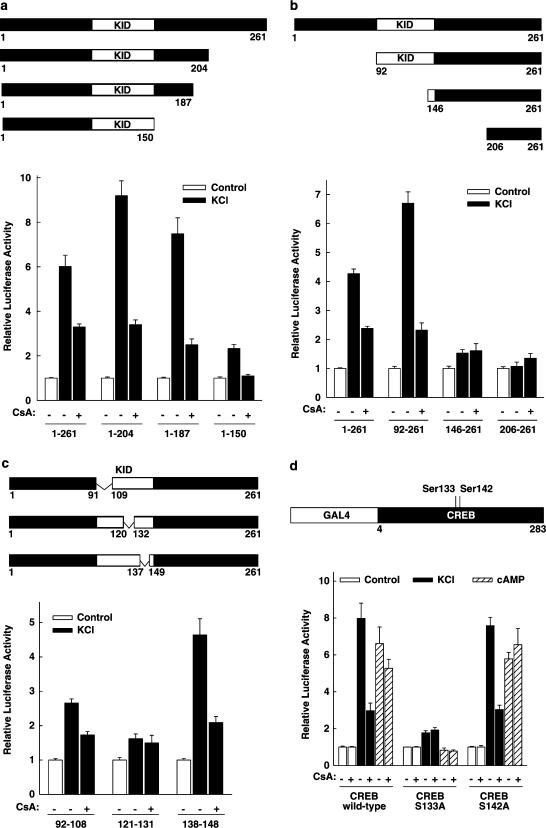
In an attempt to identify the region within the CREB transactivation domain, which confers the responsiveness to cyclosporin A after membrane depolarization, GAL4 fusion proteins containing C- and N-terminal deletions of the CREB transactivation domain were transfected together with the luciferase reporter gene. As shown in Figure 2a, membrane depolarization enhanced the transcriptional activities of the three C-terminally deleted fragments of the CREB transactivation domain; when compared to the entire transactivation domain (aa 1–261, six-fold stimulation), the stimulation was nine-fold (aa 1–204), seven-fold (aa 1–187), and two-fold (aa 1–150). Cyclosporin A inhibited the depolarization-induced activity of each fusion protein (Figure 2a). When compared to the entire CREB transactivation domain, the membrane depolarization-induced enhancement of the CREB fragment in which the aa from 205 to 261 are deleted was somewhat higher. This region is known to bind to the TBP-associated factor 110 (Ferreri et al., 1994). The deletion of the N-terminal 91 aa resulted in a slightly enhanced stimulation of transcriptional activity by membrane depolarization (seven-fold) (Figure 2b), which was fully responsive to cyclosporin A (Figure 2b). The transcriptional activities of further N-terminal CREB deletions were no longer activated by membrane depolarization (Figure 2b). In order to examine whether regions within the kinase-inducible domain (KID) of CREB confer the responsiveness to cyclosporin A treatment after membrane depolarization, the effect of internal deletions within the KID was investigated. As shown in Figure 2c, the internal deletion of aa 92–108 or aa 138–148 did not abolish the CREB response to membrane depolarization and cyclosporin A. A CREB deletion mutant lacking aa 121–131 was no longer activated by membrane depolarization and the addition of cyclosporin A had no effect (Figure 2c). The aa 121–131 of CREB play a major role in the interaction of CREB with CBP. The solution structure of the CREB–CBP complex shows that the KID of CREB undergoes a coil to helix folding transition upon binding to CBP (Radhakrishnan et al., 1997). The aa 121–131 form the amphipathic helix αB of KID that mediates most of the interactions with CBP (Parker et al., 1996; Radhakrishnan et al., 1997). In GH3 cells, the phosphorylation of Ser142 (in CREB-341) by the calcium/calmodulin-dependent protein kinase II has been shown to diminish the transactivating effect of the phosphorylation on Ser133 (Sun et al., 1994). Thus, inhibition of calcineurin activity resulting in a persistent phosphorylation of CREB on Ser142 might be one mechanism how cyclosporin A could decrease CREB transcriptional activity. In order to test this hypothesis, the effect of cyclosporin A on the transcriptional activity of the CREB Ser142Ala mutant was investigated. As shown in Figure 2d, the transcriptional activity of the transactivation domain of wild-type CREB-341 was enhanced by membrane depolarization and by forskolin (eight- and seven-fold, respectively). The addition of cyclosporin A decreased only membrane depolarization-induced transcriptional activity (inhibition by 70%), demonstrating that the inhibition of the membrane depolarization-induced transcriptional activity of the CREB transactivation domain by cyclosporin A is independent of the CREB splice variant. The transcriptional activity of the CREB Ser133Ala mutant was only marginally enhanced by membrane depolarization, and cyclosporin A had no effect (Figure 2d). The transcriptional activity conferred by the CREB Ser142Ala mutant was enhanced by membrane depolarization or cAMP to the same extent as the one conferred by wild-type CREB, and the membrane depolarization-induced transcription was still inhibited by cyclosporin A (Figure 2d). Thus, the inhibitory effect of cyclosporin A after membrane depolarization does not depend on the phosphorylation of Ser142. Taken together, these results provide no evidence for a distinct cyclosporin A-sensitive domain of CREB and suggest that cyclosporin A does not inhibit depolarization-induced CREB activity by targeting CREB itself.
Figure 2.
Effect of deletions or a mutation within the CREB transactivation domain on the inhibition by cyclosporin A of membrane depolarization-induced CREB transcriptional activity. (a) C-terminal deletion analysis. The expression vectors for the indicated GAL4-CREB fusion proteins were transfected into HIT cells together with the luciferase reporter gene controlled by five copies of the GAL4-binding site (5xGal4E1BLuc). The white box represents the kinase-inducible domain of CREB (KID), the numbers indicate the aa present (referring to CREB-327). Cells were treated as indicated with KCl (45 mM) and cyclosporin A 5 μM (CsA). Luciferase activity is expressed relative to the mean value in each experiment of the activity measured in the respective control. Values are means±s.e.m. (n=6). (b) N-terminal deletion analysis. The expression vectors for the indicated GAL4-CREB fusion proteins were transfected together with the luciferase reporter gene controlled by five copies of the GAL4-binding site (5xGal4E1BLuc). The white box represents the KID, the numbers indicate the aa present (referring to CREB-327). Cells were treated as indicated with KCl (45 mM) and cyclosporin A 5 μM (CsA). Luciferase activity is expressed relative to the mean value in each experiment of the activity measured in the respective control. Values are means±s.e.m. (n=6). (c) Analysis of internal deletions within the KID. The expression vectors for the indicated GAL4-CREB fusion proteins were transfected together with the luciferase reporter gene controlled by five copies of the GAL4-binding site (5xGal4E1BLuc). The white box represents the KID, the numbers indicate the aa present (referring to CREB-327). Δ indicates the amino acids internally deleted. Cells were treated as indicated with KCl (45 mM) and cyclosporin A 5 μM (CsA). Luciferase activity is expressed relative to the mean value in each experiment of the activity measured in the respective control. Values are means±s.e.m. (n=6). (d) Ser142 of CREB-341 is not required for cyclosporin A sensitivity of the CREB transactivation domain. Expression vectors for the GAL4 DNA-binding domain fused to the transactivation domain of CREB-341 either wild type (CREB wild-type) or bearing a Ser133Ala mutation (CREB S133A) or a Ser142Ala mutation (CREB S142A) were transfected together with the luciferase reporter gene under control of five copies of the GAL4-binding site (5xGal4E1BLuc). Ser133 of CREB-341 corresponds to Ser119 of CREB-327. Cells were treated as indicated with KCl (45 mM), forskolin 10 μM (cAMP), and cyclosporin A 5 μM (CsA). Luciferase activity is expressed relative to the mean value in each experiment of the activity measured in the respective control. Values are means±s.e.m. (n=6).
Effect of cyclosporin A on the interaction between CREB and CBP
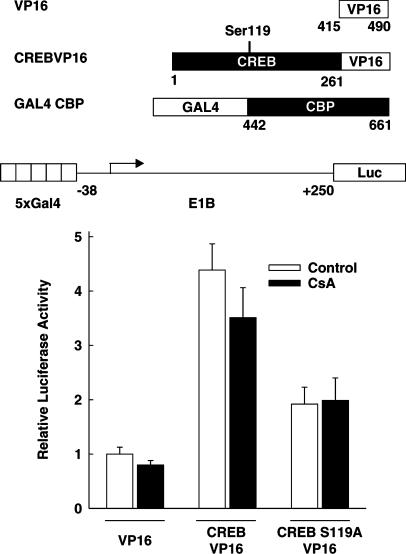
To investigate whether cyclosporin A disturbs in vivo the interaction between CREB and CBP, a mammalian two-hybrid system was employed. This approach has been successfully used to study the interaction of CBP with c-Fos (Bannister & Kouzarides, 1995) and CREB (Sun & Maurer, 1995). The luciferase reporter gene under the control of five GAL4-binding sites (5xGal4E1BLuc) was cotransfected with an expression vector encoding the DNA-binding domain of GAL4 fused to the CREB interaction domain of CBP from aa 442 to 661 (GAL4-CBP) (Figure 3); in addition, HIT cells were transfected with another expression vector, which encoded either the transactivation domain of the viral protein VP16 alone (VP16), a strong transcriptional activator, or the CREB transactivation domain fused to VP16 (CREB-VP16) (Figure 3). As shown in Figure 3, when compared to the cotransfection of VP16, the cotransfection of the expression vector for CREB-VP16 resulted in a four-fold increase of the transcriptional activity indicating the interaction between CBP and CREB. This interaction depended on the phosphorylation of CREB on Ser119, since cotransfection of a CREB-VP16 mutant, containing a Ser119 to Ala mutation of CREB (CREB-S119A-VP16), with GAL4-CBP resulted in only a very slight increase in transcriptional activity (Figure 3). Cyclosporin A did not decrease the transcriptional activity conferred by CREB-VP16 (Figure 3). The transcriptional activity of VP16 fused to the DNA-binding domain of GAL4 (GAL4-VP16) was also not decreased by cyclosporin A treatment (data not shown). These data suggest that the inhibition of CREB-dependent transcription by cyclosporin A might not be due to a decrease in the interaction between phosphorylated CREB and CBP.
Figure 3.
Effect of cyclosporin A on the interaction between CREB and its coactivator CBP as indicated by a mammalian two-hybrid assay. The luciferase reporter gene under the control of five GAL4-binding sites (5xGal4E1BLuc; 1 μg per 6-cm dish) was transfected into HIT cells together with an expression vector encoding the DNA-binding domain of GAL4 fused to the CREB interaction domain of CBP (aa 442–661) (GAL4-CBP; 1 μg per 6-cm dish). In addition, the cells were transfected with one of the following three expression vectors (2 μg per 6-cm dish): an expression vector coding for the transactivation domain of the viral protein VP16 (VP16), an expression vector encoding the CBP interaction domain of CREB-327 fused to the transactivation domain of VP16 (CREB-VP16), or an expression vector encoding a mutant CREB-VP16 protein with Ser119 of CREB mutated to Ala (CREB-S119A-VP16). Cells were treated with cyclosporin A 5 μM (CsA) as indicated. Luciferase activity is expressed relative to the activity measured in each experiment of the VP16 control. Values are means±s.e.m. (n=9).
Effect of cyclosporin A on membrane depolarization-induced CBP transcriptional activity
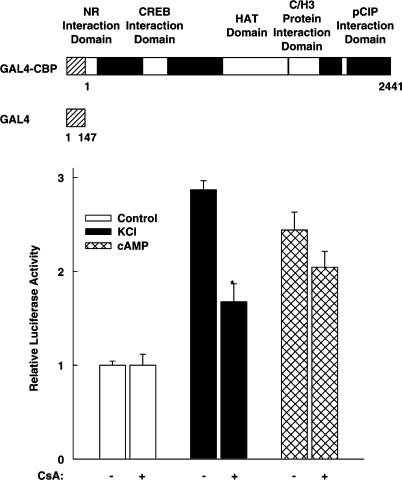
To investigate whether cyclosporin A inhibits the transcriptional activity of CBP itself, the luciferase reporter gene under the control of five GAL4-binding sites (5xGal4E1BLuc) was transfected into HIT cells together with an expression vector for a GAL4-CBP full-length fusion protein (Figure 4). As shown in Figure 4, membrane depolarization as well as cAMP stimulated the transcriptional activity of GAL4-CBP. These data confirm in this pancreatic islet β-cell line that the coactivator itself is regulated by the same stimuli that are known to enhance CREB transcriptional activity (Kwok et al., 1994; Habener et al., 1995; Chawla et al., 1998; Mayr & Montminy, 2001). The addition of cyclosporin A did reduce neither the basal nor the cAMP-induced transcriptional activity of CBP (Figure 4). In contrast, cyclosporin A inhibited CBP-directed transcriptional activity after membrane depolarization by 63% (P <0.01 versus without cyclosporin A) (Figure 4). Thus, these data suggest that cyclosporin A inhibits depolarization-induced CREB activity at the CREB coactivator level.
Figure 4.
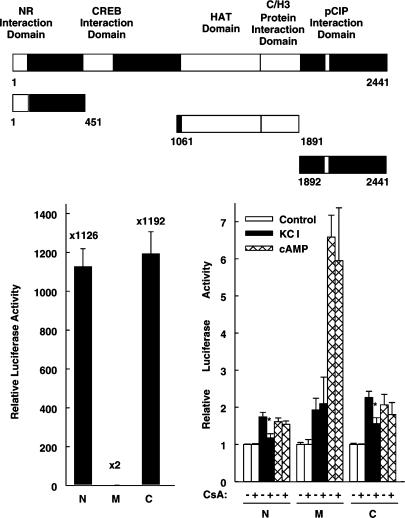
Cyclosporin A inhibits the transcriptional activity of the CREB coactivator CBP after membrane depolarization. The luciferase reporter gene under the control of five GAL4-binding sites (5xGal4E1BLuc) was transfected into HIT cells together with an expression vector for GAL4 fused to full-length CBP (GAL4-CBP). In the GAL4-CBP schematic diagram, the number 1 denotes the first aa of the CBP protein. The white boxes within CBP indicate various CBP regions. NR, nuclear receptor; HAT, histone acetyltransferase; C/H3, zinc finger. Cells were treated as indicated with KCl (45 mM), forskolin 10 μM (cAMP), and cyclosporin A 5 μM (CsA). Luciferase activity is expressed relative to the mean value in each experiment of the activity measured in the respective control. *P<0.01 versus without CsA, Student's t-test. Values are means±s.e.m. (n=6).
CBP is a large modular protein containing various interaction domains with other transcription factors and cofactors (Figure 4, upper panel). In addition, it has an intrinsic histone acetyltransferase activity (Figure 4). In order to identify a region within the CBP molecule that is sensitive to cyclosporin A treatment after membrane depolarization, different CBP domains were fused to the DNA-binding domain of GAL4.
The transcriptional activity of the KIX domain, representing the minimal CREB interaction domain of CBP (Parker et al., 1996) (GAL4-KIX), was stimulated by membrane depolarization and cAMP; after both stimuli it was decreased by cyclosporin A (data not shown). The overexpression of a dominant-negative CREB protein bearing a Ser133 to Ala mutation decreased the cAMP- and membrane depolarization-induced transcriptional activity of GAL4-KIX by 67% (data not shown), thereby indicating that the transcriptional activity of KIX is mostly due to the recruitment of endogenous CREB whose activity after recruitment is inhibited by cyclosporin A. Thus, KIX cannot be viewed as an intrinsically cyclosporin A-sensitive domain of CBP.
Both, the N-terminus of CBP and its C-terminus (aa 1–451 and aa 1892–2441, respectively) exhibited high basal transcriptional activity (Figure 5, left panel), which was further stimulated by membrane depolarization and cAMP (Figure 5, right panel). The high basal activity of the CBP N- and C-terminus (Figure 5, left panel) confirms in HIT cells what has been described previously in CV1 and HeLa cells (Kurokawa et al., 1998). Compared with the respective controls (100%), the basal activity of the N-terminus and the C-terminus of CBP was 107±9 and 85±5%, respectively (n=6, each), after overexpression of the dominant-negative CREB protein (CREB Ser133Ala). In the presence of the dominant-negative CREB protein, membrane depolarization induced the transcriptional activity of the CBP N- and C-terminus to 196±11 and 225±3%, respectively (n=6), and thus to levels similar to those in the absence of the dominant-negative CREB protein (Figure 5, right panel and data not shown). CREB Ser133Ala also had no effect on cAMP-induced activity (data not shown). Thus, these data suggest that both, the high basal activity and the inducible activity of the N- and C-terminus of CBP, does not depend on the recruitment of endogenous CREB. Cyclosporin A decreased the transcriptional activity of the N- and C-terminus of CBP induced by membrane depolarization without affecting basal or cAMP-induced transcription (Figure 5, right panel). The middle domain of CBP (aa 1061–1891), which includes the domain exhibiting histone acetyltransferase activity, conferred very low basal transcriptional activity that was enhanced by membrane depolarization and cAMP (Figure 5) but never reached the level conferred by the N- or the C-terminus of CBP. Cyclosporin A did not inhibit the transcriptional activity of the CBP middle domain (Figure 5). Thus, the CBP N- and C-terminus confer sensitivity to cyclosporin A treatment after membrane depolarization.
Figure 5.
The N- and the C-terminus of CBP confer sensitivity to cyclosporin A treatment after membrane depolarization. The upper panel shows a scheme of full-length CBP, the N-terminus (N), the middle domain (M), and the C-terminus of CBP (C). The white boxes within CBP indicate various CBP regions. NR, nuclear receptor; HAT, histone acetyltransferase; C/H3, zinc finger 3. The luciferase reporter gene under the control of five GAL4-binding sites (5xGal4E1BLuc) was transfected into HIT cells together with expression vectors for GAL4 fused to the three CBP regions depicted in the upper panel. Left panel, basal transcriptional activities of the N-terminus (N), the middle domain (M), and the C-terminus (C) of CBP. Luciferase activity is expressed relative to the mean value measured in each experiment of the GAL4 DNA-binding domain. Values are means±s.e.m. (n=6). Right panel, cells were treated as indicated with KCl (45 mM), forskolin 10 μM (cAMP), and cyclosporin A 5 μM (CsA). Luciferase activity is expressed relative to the mean value in each experiment of the activity measured in the respective control. *P<0.01 versus without CsA, Student's t-test. Values are means±s.e.m. (n=6).
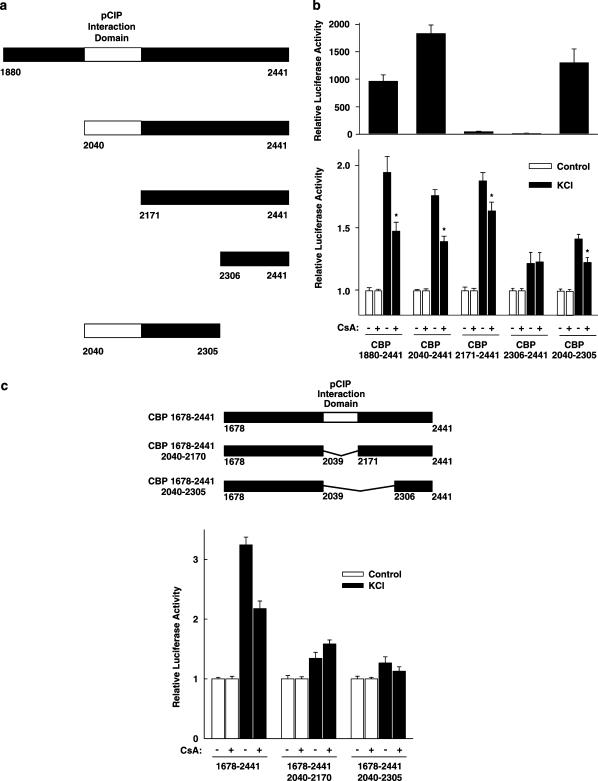
In order to localize more closely a region within the C-terminus of CBP that is sensitive to cyclosporin A after membrane depolarization, various expression vectors encoding smaller domains of the CBP C-terminus were constructed (Figure 6a and c). Upon successive N-terminal deletion, basal activity was largely lost when the domain from aa 2040 to 2170 was removed (Figure 6b, upper panel). This domain represents the minimal interaction domain of CBP with the cofactor pCIP (Torchia et al., 1997). Nevertheless, the low transcriptional activity of CBP (2171–2441) was still enhanced by membrane depolarization and somewhat inhibited by cyclosporin A (Figure 6b, lower panel). CBP (2040–2305) was the smallest CBP C-terminal fragment tested that was sufficient to confer full basal activity, membrane depolarization responsiveness, and cyclosporin A sensitivity (Figure 6b). To study whether this fragment is also required for cyclosporin A sensitivity of the CBP C-terminus, expression vectors coding for the CBP C-terminus from aa 1678 to 2441 with internal deletions were constructed (Figure 6c). As shown in Figure 6c, the internal deletion of aa 2040–2305 and also the internal deletion of only aa 2040–2170 abolished membrane depolarization responsiveness and cyclosporin A sensitivity. The internal deletion of aa 2040–2170 and aa 2040–2305 decreased basal activity to 46±6 and 32±3% of wild-type CBP C-terminus activity, respectively (n=6, each). Taken together, these data suggest that aa 2040–2170, representing the minimal interaction domain of CBP with the cofactor pCIP, are required for membrane depolarization responsiveness and cyclosporin A sensitivity of the CBP C-terminus.
Figure 6.
Mapping of a region within the CBP C-terminus conferring cyclosporin A sensitivity after membrane depolarization. (a) Scheme of the C-terminal CBP regions that were fused to the DNA-binding domain of GAL4. The white box indicates the minimal interaction domain of CBP with the cofactor pCIP ranging from aa 2040 to 2170. The numbers indicate the aa present. (b) The luciferase reporter gene under the control of five copies of the GAL4-binding site (5xGal4E1BLuc) was transfected into HIT cells together with expression vectors for the GAL4 DNA-binding domain fused to the indicated CBP C-terminal regions. Upper panel, basal transcriptional activity. Luciferase activity is expressed relative to the mean value measured in each experiment of the GAL4 DNA-binding domain. Values are means±s.e.m. (n=12). Lower panel, cells were treated with KCl (45 mM) and cyclosporin A 5 μM (CsA) as indicated. Luciferase activity is expressed relative to the mean value in each experiment of the activity measured in the respective control. *P<0.01 versus without CsA, Student's t-test. Values are means±s.e.m. (n=12). (c) Internal deletions within the CBP C-terminus. A luciferase reporter gene under the control of five copies of the GAL4-binding site (5xGal4E1BLuc) was transfected into HIT cells together with expression vectors for the GAL4 DNA-binding domain fused to the CBP C-terminus (aa 1678–2441), containing internal deletions as depicted in the upper panel. The numbers indicate the aa present, the white box represents the pCIP interaction domain of CBP (aa 2040–2170); Δ indicates the amino acids internally deleted. Cells were treated with KCl (45 mM) and cyclosporin A 5 μM (CsA) as indicated. Luciferase activity is expressed relative to the mean value in each experiment of the activity measured in the respective control. Values are means±s.e.m. (n=6).
Membrane depolarization-induced transcriptional activity of the CBP C-terminus depends on calcineurin phosphatase activity
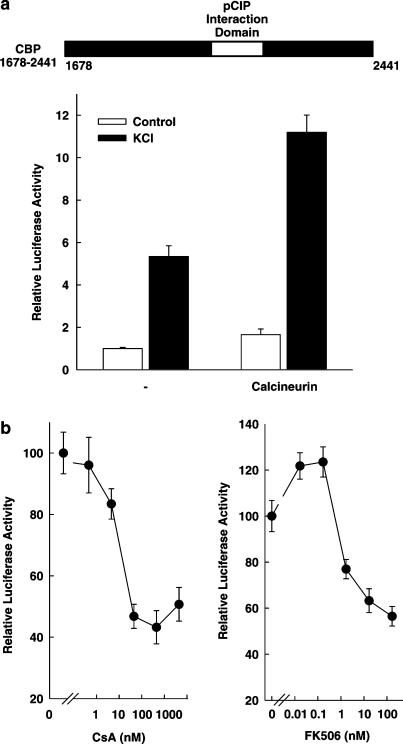
The fact that cyclosporin A (5 μM) inhibits CBP transcriptional activity after membrane depolarization suggests that calcineurin phosphatase activity contributes to the stimulation by membrane depolarization of CBP transcriptional activity. This suggested importance of calcineurin for CBP activity was further investigated. The overexpression of calcineurin A and B subunits enhanced the transcriptional activity of the CBP C-terminus (from aa 1678 to 2441) both under basal conditions and after stimulation by membrane depolarization (Figure 7a). Furthermore, the membrane depolarization-induced transcriptional activity of the CBP C-terminus was inhibited by both structurally distinct drugs, cyclosporin A and tacrolimus, in a concentration-dependent manner with IC50 values of 10 and 1 nM, respectively (Figure 7b). Cyclosporin A and tacrolimus have been shown before to inhibit calcineurin phosphatase activity at these concentrations in the pancreatic islet β-cell line (Schwaninger et al., 1995). Taken together, these data indicate that the membrane depolarization-induced transcriptional activity of the CREB coactivator CBP depends on calcineurin phosphatase activity. At the same time, these data suggest that the immunosuppressive drugs cyclosporin A and tacrolimus inhibit the activity of the transcription factor CREB at the coactivator level through inhibition of calcineurin activity.
Figure 7.
Calcineurin mediates part of the membrane depolarization-induced transcriptional activity of the CBP C-terminus. (a) The overexpression of calcineurin enhances CBP C-terminus-dependent transcription. The luciferase reporter gene under control of five copies the GAL4-binding site (5xGal4E1BLuc) was transfected into HIT cells together with an expression vector for the GAL4 DNA-binding domain fused to the CBP C-terminus (aa 1678–2441) and expression vectors for calcineurin subunits A and B. The white box indicates the pCIP interaction domain of CBP (aa 2040–2170). Cells were treated with KCl (45 mM) as indicated. Luciferase activity is expressed relative to the mean value in each experiment of the activity measured in the control without calcineurin. Values are means±s.e.m. (n=6). (b) Cyclosporin A and tacrolimus inhibit the transcriptional activity of the CBP C-terminus after membrane depolarization with high potency. The luciferase reporter gene under control of five copies the GAL4-binding site (5xGal4E1BLuc) was transfected together with an expression vector for the GAL4 DNA-binding domain fused to the CBP C-terminus (aa 1678–2441). Cells were stimulated by membrane depolarization (KCl 45 mM) and treated with increasing concentrations of cyclosporin A (CsA) or tacrolimus (FK506). Luciferase activity is expressed relative to the mean value in each experiment of the stimulus-induced activity in controls (no CsA, no FK506). Values are means±s.e.m. (n=6).
Discussion
Cyclosporin A and tacrolimus interfere with mechanisms of gene regulation as part of their immunosuppressive and undesired effects (Ho et al., 1996). NFAT is a substrate of calcineurin phosphatase (Ho et al., 1996). Thus, through inhibition of calcineurin, cyclosporin A and tacrolimus directly target NFAT (Ho et al., 1996). In contrast, the results of the present study suggest that, through inhibition of calcineurin, cyclosporin A and tacrolimus do not target CREB directly but rather inhibit CREB transcriptional activity at the coactivator level.
Both immunosuppressive drugs have been shown to block CRE/CREB-directed transcription after membrane depolarization or other stimuli in various cell lines through inhibition of calcineurin phosphatase activity without decreasing the phosphorylation of CREB on Ser119 (in CREB-327, corresponding to Ser133 in CREB-341) (Schwaninger et al., 1993a, 1993b; 1995; Krüger et al., 1997). This is consistent with several lines of evidence that have clearly demonstrated that the phosphorylation of CREB on Ser119 is required but not sufficient for CREB transcriptional activity (Sun et al., 1994; Sun & Maurer, 1995; Parker et al., 1996; Mayr et al., 2001; Wu & McMurray, 2001). Consequently, several signalling pathways have been shown to regulate the transcriptional activity of CREB phosphorylated on Ser119 (Fiol et al., 1994; Sun et al., 1994; Parker et al., 1996; 1998; Wu & McMurray, 2001; Gau et al., 2002; Kornhauser et al., 2002). These regulatory mechanisms increase the complexity of CREB regulation and thus allow to impart a greater degree of specificity to target gene activation via CREB. These signalling pathways influence different steps in phosphoCREB-mediated transactivation, acting either on CREB directly, on CREB–CBP interaction, or on the coactivator CBP itself. However, it was unknown at what step in CREB-mediated transcription cyclosporin A and tacrolimus interfere.
Consistent with signalling directly on CREB, previous studies have identified sites on CREB in addition to Ser119/133 that can be phosphorylated in vitro or by overexpression of protein kinases in cells. These examples include (in CREB-341) Ser129 phosphorylation by glycogen synthase kinase-3 (Fiol et al., 1994), Ser142 phosphorylation by calcium/calmodulin-dependent protein kinase II (CaMKII) (Sun et al., 1994), and phosphorylation of either Ser142 or Ser143 by casein kinase II (Parker et al., 1998). At least some of these phosphorylations like Ser142 phosphorylation also occur in vivo in response to external stimuli such as depolarization-induced calcium influx (Gau et al., 2002; Kornhauser et al., 2002). Although the functional consequences of Ser142 phosphorylation seem to depend on the specific conditions (Gau et al., 2002: Kornhauser et al., 2002), Ser133 phosphorylation of CREB by CaMKII fails to promote target gene activation because of secondary CaMKII-dependent phosphorylation of CREB at Ser142 (Sun et al., 1994; Wu & McMurray, 2001), indicating an inhibitory role of Ser142 phosphorylation. In the present study, the mutation of Ser142 of CREB to Ala did not alter depolarization-induced CREB activity, suggesting that phosphorylation of Ser142 does not regulate CREB transcriptional activity after membrane depolarization in the pancreatic islet β-cell line. In addition, cyclosporin A inhibited depolarization-induced CREB activity also after mutation of Ser142. This indicates that cyclosporin A does not exert its inhibitory action through Ser142 of CREB. Furthermore, cyclosporin A inhibited depolarization-induced activity of GAL4-CREB proteins that carried extensive deletions within the CREB transactivation domain. These data provide no evidence for the existence of a distinct cyclosporin A-sensitive domain of CREB and thereby suggest that the immunosuppressive drugs through inhibition of calcineurin may not act on CREB itself but at a different level in CREB-mediated transcription.
Several signals have been shown to regulate the interaction between phosphoserine-133-CREB and its coactivator CBP. The solution structure of the CREB–CBP complex, using relevant interaction domains referred to as KID and KIX, respectively, reveals that phosphoSer133 promotes complex formation by means of ion-pair and hydrogen-bond interactions with residues in KIX (Radhakrishnan et al., 1997); the Ser133 phosphate also stabilizes the formation of an amphipathic helix in KID that associates with a shallow hydrophobic groove in KIX (Radhakrishnan et al., 1997). The solution structure thus demonstrates that Ser133 phosphorylation of CREB is both necessary and sufficient for complex formation (Radhakrishnan et al., 1997). Nevertheless, mitogen and stress signals are far less effective than cAMP in stimulating CREB–CBP complex formation even though they induce comparable levels of Ser133 phosphorylation (Mayr et al., 2001). This inhibition of phosphoSer133-CREB–CBP interaction by mitogen and stress signals does not seem to be secondary to covalent modifications of either CREB or CBP but may be caused by a nuclear inhibitory protein (Mayr et al., 2001). The immunosuppressants appear to inhibit CRE/CREB activity through a different mechanism, because in the present study cyclosporin A did not inhibit the interaction between phosphoCREB and CBP as indicated by a two-hybrid assay. When taken together, the results of the present study suggest that the immunosuppressants may block CREB activity by inhibiting CBP function, thus acting at the CREB coactivator level.
It was thought initially that coactivators like CBP confer constitutive transcriptional activity to a promoter. Meanwhile, it became clear that signalling pathways regulate not only the recruitment of CBP but also target CBP directly to stimulate or inhibit CBP transcriptional activity (Kwok et al., 1994; Chawla et al., 1998). Consistent with previous studies in the mouse pituitary cell line AtT20 (Chawla et al., 1998), in cortical neurons (Hu et al., 1999), and in hippocampal neurons (Impey et al., 2002), the present study shows that membrane depolarization stimulates CBP transcriptional activity in a pancreatic islet β-cell line, with responsive domains located both in the N-terminal and C-terminal regions of CBP. The mechanisms involved are incompletely understood but may involve CaMKIV activity and phosphorylation of Ser301 of CBP (Impey et al., 2002). The present study now suggests that calcineurin phosphatase activity is also involved in depolarization-induced CBP transactivation. Since depolarization-induced CBP C-terminus transcriptional activity is enhanced by overexpression of calcineurin and is inhibited by both cyclosporin A and tacrolimus with potencies similar to their potency to inhibit calcineurin, a model emerges whereby membrane depolarization results in the phosphorylation of CREB on Ser119 (presumably by nuclear CaMKIV) leading to the recruitment of the coactivator CBP whose transcriptional activity itself is enhanced by CaMKs and by calcineurin. This model implies that cyclosporin A and tacrolimus inhibit membrane depolarization-induced CRE/CREB transcriptional activity through calcineurin at the coactivator level by inhibiting CBP transactivation.
An external stimulus that causes membrane depolarization in pancreatic islet β cells is glucose, which in this way increases calcineurin phosphatase activity and insulin gene transcription (Oetjen et al., 2003a, 2003b). The human insulin gene carries four copies of the CRE and is regulated by CREB (Oetjen et al., 2003a, 2003b). On the other hand, in electrically nonexcitable Jurkat T cells, the transcriptional activity of CREB (Krüger et al., 1997) and CBP (Riggins & Clipstone, 2001) is enhanced by T-cell activation signals in a tacrolimus-sensitive manner. Normal T-cell activation depends on CREB (Barton et al., 1996). Thus, the results of the present study suggest that inhibition of CREB transcriptional activity at the CBP coactivator level may contribute to both the desired immunosuppressive and the undesired diabetogenic effects of cyclosporin A and tacrolimus.
Acknowledgments
The present study was supported by a grant from the Deutsche Forschungsgemeinschaft, SFB402/A3. We appreciate these generous gifts: expression vectors for various GAL4-CREB fusion proteins from Dr Joel F. Habener, Boston, MA, U.S.A., from Dr Richard A. Maurer, Portland, OR, U.S.A., and from Dr Ourania M. Andrisani, West Lafayette, IN, U.S.A.; expression vectors for various GAL4-CBP fusion proteins from Dr Richard H. Goodman, Portland, OR, U.S.A., and from Dr Christopher Glass, San Diego, CA, U.S.A.; expression vector for VP16 from Dr Andrew Bannister, Cambridge, U.K.; expression vectors for calcineurin subunits A and B from Dr Gerald R. Crabtree, Stanford, CA, U.S.A. We thank Dr Ulrike Böer for critically reading the manuscript.
Abbreviations
- aa
amino acid(s)
- CBP
CREB-binding protein
- CREB
cAMP response element-binding protein
- CRE
cAMP response element
- KID
kinase-inducible domain
- NFAT
nuclear factor of activated T cells
- VP16
viral protein 16
References
- BANNISTER A.J., KOUZARIDES T. CBP-induced stimulation of c-Fos activity is abrogated by E1A. EMBO J. 1995;14:4758–4762. doi: 10.1002/j.1460-2075.1995.tb00157.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- BARTON K., MUTHASAMY N., CHANYANGAM M., FISCHER C., CLENDENIN C., LEIDEN J.M. Defective thymocyte proliferation and IL-2 production in transgenic mice expressing a dominant negative form of CREB. Nature (London) 1996;379:81–85. doi: 10.1038/379081a0. [DOI] [PubMed] [Google Scholar]
- CHAWLA S., HARDINGHAM G.E., QUINN D.R., BADING H. CBP: a signal-regulated transcriptional coactivator controlled by nuclear calcium and CaM kinase IV. Science. 1998;281:1505–1509. doi: 10.1126/science.281.5382.1505. [DOI] [PubMed] [Google Scholar]
- CHRIVIA J.C., KWOK R.P.S., LAMB N., HAGIWARA M., MONTMINY M.R., GOODMAN R.H. Phosphorylated CREB binds specifically to the nuclear protein CBP. Nature (London) 1993;365:855–859. doi: 10.1038/365855a0. [DOI] [PubMed] [Google Scholar]
- CLIPSTONE N.A., CRABTREE G.R. Identification of calcineurin as a key signalling enzyme in T lymphocycte activation. Nature (London) 1992;357:695–697. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- FERRERI K., GILL G., MONTMINY M. The cAMP-regulated transcription factor CREB interacts with a component of the TFIID complex. Proc. Natl. Acad. Sci. U.S.A. 1994;91:1210–1213. doi: 10.1073/pnas.91.4.1210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FIOL C.J., WILLIAMS J.S., CHOU C.H., WANG Q.M., ROACH P.J., ANDISANI O.M. A secondary phosphorylation of CREB341 at Ser129 is required for the cAMP-mediated control of gene expression. J. Biol. Chem. 1994;269:32187–32193. [PubMed] [Google Scholar]
- GAU D., LEMBERGER T., VON GALL C., KRETZ O., MINH N.L., GASS P., SCHMID W., SCHIBLER U., KORF H.W., SCHÜTZ G. Phosphorylation of CREB Ser142 regulates light-induced phase shifts of the circadian clock. Neuron. 2002;34:245–253. doi: 10.1016/s0896-6273(02)00656-6. [DOI] [PubMed] [Google Scholar]
- HABENER J.F., MILLER C.P., VALLEJO M. cAMP-dependent regulation of gene transcription by cAMP response element-binding protein and cAMP response element modulator. Vitam. Horm. 1995;51:1–57. doi: 10.1016/s0083-6729(08)61037-7. [DOI] [PubMed] [Google Scholar]
- HERZIG S., LONG F., JHALA U.S., HEDRICK S., QUINN R., BAUER A., RUDOLPH D., SCHÜTZ G., YOON C., PUIGSERVER P., SPIEGELMAN B., MONTMINY M. CREB regulates hepatic gluconeogenesis through the coactivator PGC-1. Nature (London) 2001;413:179–183. doi: 10.1038/35093131. [DOI] [PubMed] [Google Scholar]
- HO S., CLIPSTONE N., TIMMERMAN L., NORTHROP J., GRAEF I., FIORENTINO D., NOURSE J., CRABTREE G.R. The mechanism of action of cyclosporin A and FK506. Clin. Immunol. Immunopathol. 1996;80:S40–S45. doi: 10.1006/clin.1996.0140. [DOI] [PubMed] [Google Scholar]
- HU S.C., CHRIVIA J., GOSH A. Regulation of CBP-mediated transcription by neuronal calcium signaling. Neuron. 1999;22:799–808. doi: 10.1016/s0896-6273(00)80738-2. [DOI] [PubMed] [Google Scholar]
- IMPEY S., FONG A.L., WANG Y., CARDINAUX J.R., FASS D.M., OBRIETAN K., WAYMAN G.A., STORM D.R., SODERLING T., GOODMAN R.H. Phosphorylation of CBP mediates transcriptional activation by neural activity and CaM kinase IV. Neuron. 2002;34:235–244. doi: 10.1016/s0896-6273(02)00654-2. [DOI] [PubMed] [Google Scholar]
- KORNHAUSER J.M., COWAN C.W., SHAYWITZ A.J., DOLMETSCH R.E., GRIFFITH E.C., HU L.S., HADDAD C., XIA Z., GREENBERG M.E. CREB transcriptional activity in neurons is regulated by multiple, calcium-specific phosphorylation events. Neuron. 2002;34:221–233. doi: 10.1016/s0896-6273(02)00655-4. [DOI] [PubMed] [Google Scholar]
- KORZUS E., TORCHIA J., ROSE D.W., XU L., KUROKAWA R., MCINERNY E.M., MULLEN T.M., GLASS C.K., ROSENFELD M.G. Transcription factor-specific requirements for coactivators and their acetlytransferases functions. Science. 1998;279:703–707. doi: 10.1126/science.279.5351.703. [DOI] [PubMed] [Google Scholar]
- KRÜGER M., SCHWANINGER M., BLUME R., OETJEN E., KNEPEL W. Inhibition of CREB- and cAMP response element-mediated gene transcription by the immunosuppressive drugs cyclosporin A and FK506 in T cells. Naunyn Schmiedeberg's Arch. Pharmacol. 1997;356:433–440. doi: 10.1007/pl00005073. [DOI] [PubMed] [Google Scholar]
- KUROKAWA R., KALAFUS D., OGLIASTRO M.H., KIOUSSI C., XU L., TORCHIA J., ROSENFELD M.G., GLASS C.K. Differential use of CREB binding protein-coactivator complexes. Science. 1998;279:700–703. doi: 10.1126/science.279.5351.700. [DOI] [PubMed] [Google Scholar]
- KWOK R.P.S., LUNDBLAD J.R., CHRIVIA J.C., RICHARDS J.P., BÄCHINGER H.P., BRENNAN R.G., ROBERTS S.G.E., GREEN M.R., GOODMAN R.H. Nuclear protein CBP is a coactivator for the transcription factor CREB. Nature (London) 1994;370:223–226. doi: 10.1038/370223a0. [DOI] [PubMed] [Google Scholar]
- LAWRENCE M.C., BHATT H.S., WATTERSON J.M., EASOM R.A. Regulation of insulin gene transcription by a Ca(2+)-responsive pathway involving calcineurin and nuclear factor of activated T cells. Mol. Endocrinol. 2001;15:1758–1767. doi: 10.1210/mend.15.10.0702. [DOI] [PubMed] [Google Scholar]
- MANTAMADIOTIS T., LEMBERGER T., BLECKMANN S.C., KERN H., KRETZ O., VILLALBA A.M., TRONCHE F., KELLENDONK C., GAU D., KAPFHAMMER J., OTTO C., SCHMID W., SCHÜTZ G. Disruption of CREB function in brain leads to neurodegeneration. Nat. Gen. 2002;31:47–54. doi: 10.1038/ng882. [DOI] [PubMed] [Google Scholar]
- MARTINEZ-BALBAS M.A., BANNISTER A.J., MARTIN K., HAUS-SEUFFERT P., MEISTERERNST M., KOUZARIDES T. The acetyltransferase activity of CBP stimulates transcription. EMBO J. 1998;17:2886–2893. doi: 10.1093/emboj/17.10.2886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MAYR B., MONTMINY M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat. Rev. Mol. Cell. Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- MAYR B.M., CANETTIERI G., MONTMINY M.R. Distinct effects of cAMP and mitogenic signals on CREB-binding protein recruitment impart specificity to target gene activation via CREB. Proc. Natl. Acad. Sci. U.S.A. 2001;98:10936–10941. doi: 10.1073/pnas.191152098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- NAKAJIMA T., UCHIDA C., ANDERSON S.F., LEE C.G., HURWITZ J., PARVIN J.D., MONTMINY M. RNA helicase A mediates association of CBP with RNA polymerase II. Cell. 1997;90:1107–1112. doi: 10.1016/s0092-8674(00)80376-1. [DOI] [PubMed] [Google Scholar]
- OETJEN E., BAUN D., BEIMESCHE S., KRAUSE D., CIERNY I., BLUME R., DICKEL C., WEHNER S., KNEPEL W. Inhibition of human insulin gene transcription by the immunosuppressive drugs cyclosporin A and tacrolimus in primary, mature islets of transgenic mice. Mol. Pharmacol. 2003a;63:1289–1295. doi: 10.1124/mol.63.6.1289. [DOI] [PubMed] [Google Scholar]
- OETJEN E., GRAPENTIN D., BLUME R., SEEGER M., KRAUSE D., EGGERS A., KNEPEL W. Regulation of human insulin gene transcription by the immunosuppressive drugs cyclosporin A and tacrolimus at concentrations that inhibit calcineurin activity and involving the transcription factor CREB. Naunyn Schmiedeberg's Arch. Pharmacol. 2003b;367:227–236. doi: 10.1007/s00210-003-0694-7. [DOI] [PubMed] [Google Scholar]
- PARKER D., FERRERI K., NAKAJIMA T., LAMORTE V.J., EVANS R., KOERBER S.C., HOEGER C., MONTMINY M.R. Phosphorylation of CREB at Ser-133 induces complex formation with CREB-binding protein via a direct mechanism. Mol. Cell. Biol. 1996;16:694–703. doi: 10.1128/mcb.16.2.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- PARKER D., JHALA U.S., RADHAKRISHNAN I., YAFFE M.B., REYES C., SHULMAN A.I., CANTLEY L.C., WRIGHT P.E., MONTMINY M. Analysis of an activator : coactivator complex reveals an essential role for secondary structure in transcriptional activation. Mol. Cell. 1998;2:353–359. doi: 10.1016/s1097-2765(00)80279-8. [DOI] [PubMed] [Google Scholar]
- RADHAKRISHNAN I., PEREZ-ALVARADO G.C., PARKER D., DYSON H.J., MONTMINY M.R., WRIGHT P.E. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator : coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- RIGGINS P.S., CLIPSTONE N.A. T cell activation signals upregulate CBP-dependent transcriptional activity. Biochem. Biophys. Res. Com. 2001;281:842–850. doi: 10.1006/bbrc.2001.4461. [DOI] [PubMed] [Google Scholar]
- RUDOLPH D., TAFURI A., GASS P., HÄMMERLING G.J., ARNOLD B., SCHÜTZ G. Impaired fetal T cell development and perinatal lethality in mice lacking the cAMP response element binding protein. Proc. Natl. Acad. Sci. U.S.A. 1998;95:4481–4486. doi: 10.1073/pnas.95.8.4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SADOWSKI I., PTASHNE M. A vector for expressing GAL4(1–147) fusions in mammalian cells. Nucl. Acids Res. 1989;17:7539. doi: 10.1093/nar/17.18.7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SCHWANINGER M., BLUME R., KRÜGER M., LUX G., OETJEN E., KNEPEL W. Involvement of the Ca2+ dependent phosphatase calcineurin in gene transcription that is stimulated by cAMP through cAMP response elements. J. Biol. Chem. 1995;270:8860–8866. doi: 10.1074/jbc.270.15.8860. [DOI] [PubMed] [Google Scholar]
- SCHWANINGER M., BLUME R., OETJEN E., LUX G., KNEPEL W. Inhibition of cAMP-responsive element-mediated gene transcription by cyclosporin A and FK506 after membrane depolarization. J. Biol. Chem. 1993a;268:23111–23115. [PubMed] [Google Scholar]
- SCHWANINGER M., LUX G., BLUME R., OETJEN E., HIDAKA H., KNEPEL W. Membrane depolarization and calcium influx induce glucagon gene transcription in pancreatic islet cells through the cyclic AMP-responsive element. J. Biol. Chem. 1993b;268:5168–5177. [PubMed] [Google Scholar]
- SCREATON R.A., CONKRIGHT M.D., KATOH Y., BEST J.L., CANETTIERI G., JEFFRIES S., GUZMAN E., NIESSEN S., YATES J.R., III, TAKEMORI H., OKAMOTO M., MONTMINY M. The CREB coactivator TORC2 functions as a calcium- and cAMP-sensitive coincidence detector. Cell. 2004;119:61–74. doi: 10.1016/j.cell.2004.09.015. [DOI] [PubMed] [Google Scholar]
- SIEMANN G., BLUME R., GRAPENTIN D., OETJEN E., SCHWANINGER M., KNEPEL W. Inhibition of cyclic AMP response element-binding protein/cyclic AMP response element-mediated transcription by the immunosuppressive drugs cyclosporin A and FK506 depends on the promoter context. Mol. Pharmacol. 1999;55:1094–1100. doi: 10.1124/mol.55.6.1094. [DOI] [PubMed] [Google Scholar]
- SUN P., ENSLEN H., MYUNG P.S., MAURER R.A. Differential activation of CREB by Ca2+/calmodulin dependent protein kinase type II and IV involves phosphorylation of a site that negatively regulates activity. Genes Dev. 1994;8:2527–2539. doi: 10.1101/gad.8.21.2527. [DOI] [PubMed] [Google Scholar]
- SUN P., MAURER R.A. An inactivating point mutation demonstrates that interaction of cAMP response element binding protein (CREB) with the CREB binding protein is not sufficient for transcriptional activation. J. Biol. Chem. 1995;270:7041–7044. doi: 10.1074/jbc.270.13.7041. [DOI] [PubMed] [Google Scholar]
- TORCHIA J., ROSE D.W., INOSTROZA J., KAMEI Y., WESTIN S., GLASS C.K., ROSENFELD M.G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear receptor function. Nature (London) 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- WU X., MCMURRAY C.T. Calmodulin kinase II attenuation of gene transcription by preventing CREB dimerization and CBP binding. J. Biol. Chem. 2001;276:1735–1741. doi: 10.1074/jbc.M006727200. [DOI] [PubMed] [Google Scholar]