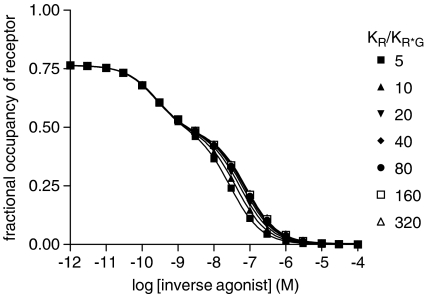
Figure 5.
Simulation of inverse agonist/[3H]agonist competition using the extended ternary complex model (Figure 1). The extended ternary complex model for two ligands was used with the following parameters: Rtot=1 × 10−7 M, Gtot=5 × 10−8 M, L=100, J=1 × 10−10 M; dissociation constants for inverse agonist: KR=1 × 10−10 M, KR*=KR*G; dissociation constants for agonist: KR=5 × 10−9 M, KR*=1 × 10−11 M, KR*G=1 × 10−11 M. KR*G was varied to give the KR/KR*G ratios shown, data were simulated in Excel ([agonist]=1 nM, varying concentrations of inverse agonist) and fitted using Prism as described in Methods section. Parameters derived from this analysis are given in Table 4.