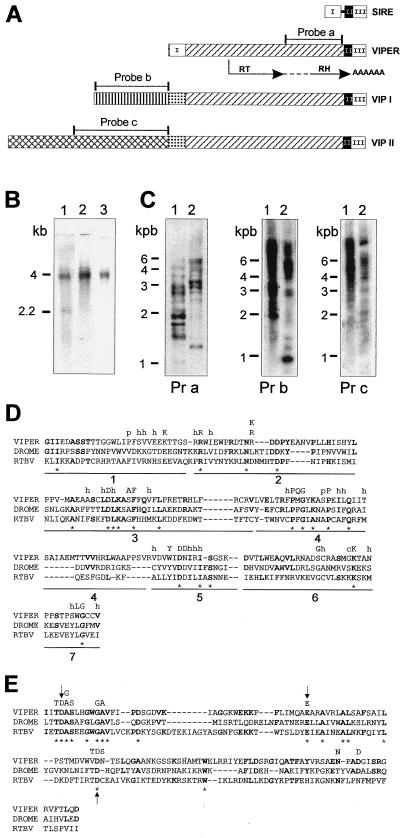
Figure 5.
(A) Schematic representation of VIPER elements. Identical boxes represent highly homologous regions. SIRE is represented with its three regions as in Fig. 2A. The connecting line between region I and region II represents the 30-bp sequence of SIRE lost in VIPER. The RT and RH domains are indicated by arrows below the diagram of VIPER; the dotted line connecting the two domains indicates the 103-residue-long tether. The position of the probes used in hybridization experiments also are indicated. (B) Northern blot analysis of T. cruzi epimastigotes poly(A)+ RNA. Lane 1, probe a; lane 2, probe b; lane 3, probe c. Sizes of the bands identified are indicated on the left. (C) Southern blot analysis of T. cruzi genomic DNA. Lane 1, EcoRI digest; lane 2, BamHI digest. Pr a, probe a; Pr b, probe b; Pr c, probe c. Sizes of the molecular DNA markers are indicated on the left. (D) Alignment of VIPER RT domains with those of Drosophila melanogaster Burdock LTR-retrotransposon, DROME (18) and rice tungro bacilliform pararetrovirus, RTBV (19). The conserved subdomains proposed by Xiong and Eickbush (17) are numbered 1–7. Amino acids conserved between VIPER and the other two sequences are shown in bold, and * denotes residues conserved in all three. The highly conserved residues in the analysis of Xiong and Eickbush (17) are indicated above the comparison at the corresponding position; h, hydrophobic residue; p, small polar residue. (E) Alignment of RH domains. Highly conserved residues in the analysis of Doolittle et al. (20) are noted above the amino acid sequence. Arrows indicate the three crucial residues referred to in the text.