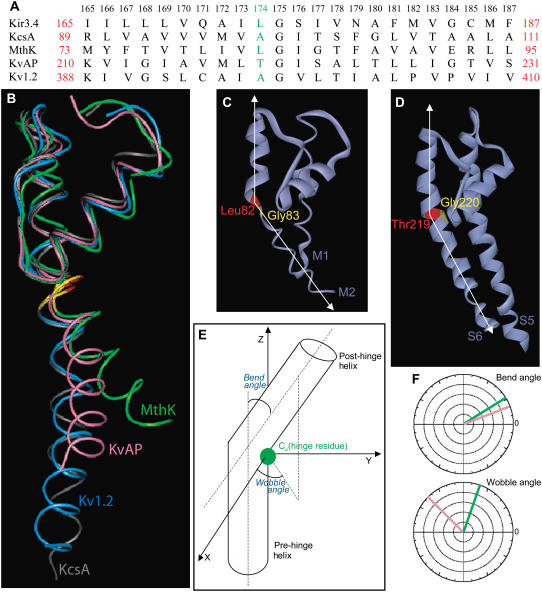
FIGURE 1.
(A) Sequence alignment between residues 165–187 of the inner helix of Kir3.4 with the corresponding residues of KcsA, MthK, KvAP, and Kv1.2. Leu-174 of Kir3.4, Ala-98 of KcsA, Leu-82 of MthK, Thr-219 of KvAP, and Ala-397 of Kv1.2 are highlighted. The numbers above the alignment refer to Kir3.4 numbers. (B) Relative orientation of the inner helix of KcsA (gray), MthK (green), KvAP (pink), and Kv1.2 (blue). (C) The TM pore domain of the open conformation of MthK (PDB code 1LNQ), showing the bending of the inner helix at Leu-82 (red), the position that precedes the central Gly-83 (yellow). (D) The TM pore domain of the open conformation of KvAP (PDB code 1ORQ), showing the bending of the inner S6 helix at Thr-219 (red), the position that precedes the central Gly-220 (yellow). (E) Diagram displaying the bend and wobble angles in a hinged helix. (F) Bend and Wobble angles of the inner helices of MthK (green) and KvAP (pink) as obtained using the program Simulaid (29).