Abstract
The melatonin rhythm-generating enzyme, arylalkylamine N-acetyltransferase (AANAT) is known to have recognizable ancient homologs in bacteria and fungi, but not in other eukaryotes. Analysis of new cDNA and genomic sequences has identified several additional homologs in other groupings. First, an AANAT homolog has been found in the genome of the cephalochordate amphioxus, representing the oldest homolog in chordates. Second, two AANAT homologs have been identified in unicellular green algae. The homologs in amphioxus, unicellular green algae, fungi and bacteria are similarly primitive in that they lack sequences found in vertebrate AANATs that are involved in regulation and that facilitate binding and catalysis. In addition, all these sequences are intronless. These features are consistent with horizontal transfer of the AANAT ancestor from bacteria to green algae, fungi and chordates. Lastly, a third AANAT gene has been found in teleost fish, suggesting that AANAT genes serve multiple functions in addition to melatonin synthesis.
Keywords: arylalkylamine N-acetyltransferase, AANAT, evolution, melatonin
1. Introduction
The best documented role of arylalkylamine N-acetyltransferase (AANAT) is in melatonin synthesis, in which it is the penultimate enzyme in the conversion of tryptophan to melatonin (Klein et al., 1997). AANAT acetylation of serotonin generates N-acetylserotonin, which is then converted to melatonin by hydroxyindole O-methyltransferase (HIOMT). Changes in the activity of AANAT control the daily rhythm in melatonin production in the pineal gland. Circulating melatonin levels are higher at night, and provide a hormonal signal of circadian and seasonal time (Arendt, 1995). Pineal AANAT activity in mammals is controlled by output from a circadian oscillator located in the suprachiasmatic nucleus. In pineal organs of lower vertebrates, AANAT is regulated by a circadian clock located in the pinealocyte (Klein et al., 1998; Iuvone et al., 2005).
The retina is the only other tissue that expresses significant levels of AANAT. Expression follows a daily rhythm, which is driven by an endogenous clock in photoreceptors cells (Cahill et al., 1991; Tosini and Menaker, 1996; Tosini, 2000; Falcón et al., 2003). AANAT functions to control melatonin synthesis in some vertebrate retinas; however, it has been proposed that it also plays a role in the retina in arylalkylamine detoxification (Coon et al., 2002). A single AANAT gene occurs in mammalian, avian and anuran genomes; fish have been reported to have two isoforms, AANAT-1 and AANAT-2, preferentially expressed in the retina and pineal gland, respectively (Bégay et al.,1998; Coon et al., 1999; Benyassi et al., 2000; Falcón et al., 2003; Zilberman-Peled et al., 2004).
Vertebrate AANATs and their ancestral homologs form a family within the GCN5-related N-acetyltransferase superfamily (GNAT superfamily; Dyda et al., 2000; Vetting et al., 2005). Members of this superfamily possess a conserved AcCoA binding domain and share a common mechanism of acetyl transfer; however, each family has markedly different substrate preference, ranging from histones to aminoglycosides. This diversification is apparent in bacteria (Vetting et al., 2005).
Analysis of the available databases has revealed that the biological distribution of AANAT is unusual in that homologs appear to occur only in the genomes of gram-positive bacteria, fungi and vertebrates, and not in other published genomes, including higher plants, nematodes and arthropods (Fig. 1; Iyer et al., 2004). Moreover, it has been argued that this pattern could have resulted from horizontal gene transfer (HGT) of the ancestor of AANAT from bacteria (Iyer et al., 2004). HGT occurs when a gene is transferred directly from one organism into the germ line of an unrelated species. This is in constrast to the conventional mechanism of vertical gene transfer whereby new genes evolve very slow as a result of mutation. HGT has the obvious feature of allowing rapid acquisition of a new capacity. Further, it needs to happen only once to have a long lasting impact on the evolution of the recipient.
Figure 1.
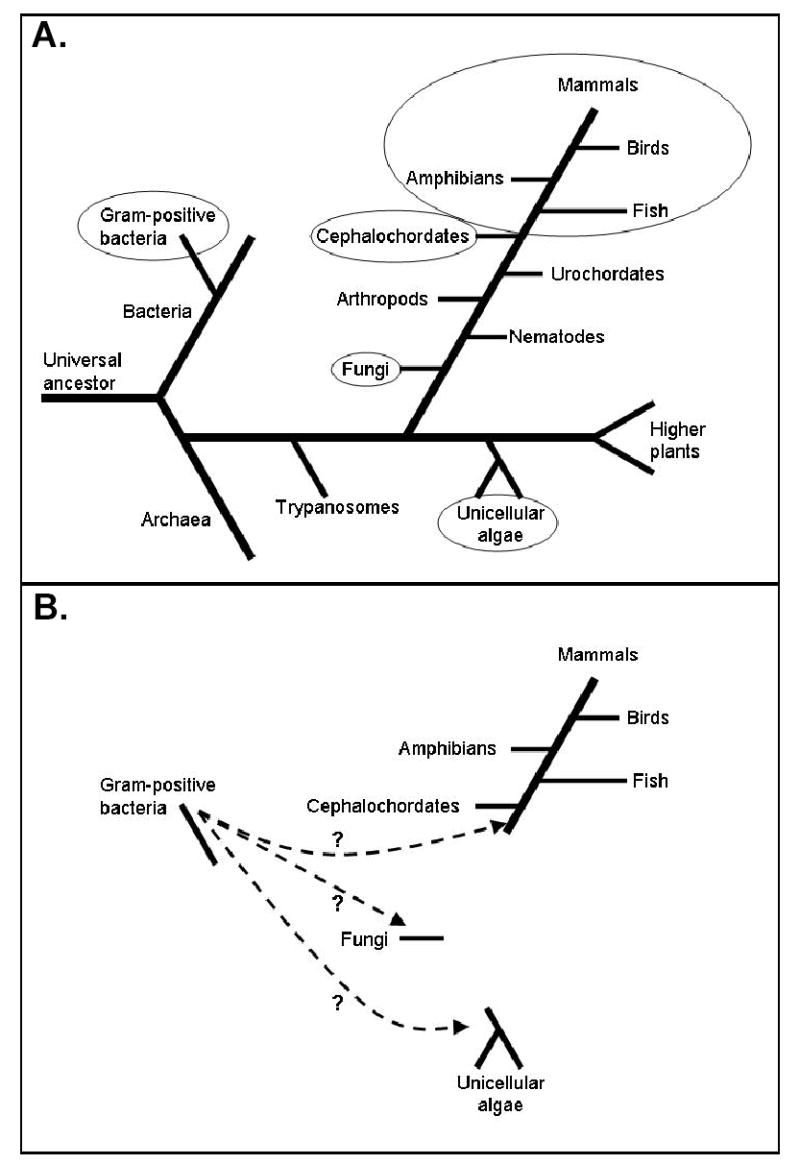
Schematic phylogenetic tree showing selective major phyla. A. Circled phyla contain AANAT homologs. Other phyla shown contain organisms whose sequenced genomes do not contain an AANAT homolog. B. Hypothetical scheme showing that an ancestral AANAT homolog may have been acquired by horizontal gene transfer at least three times.
AANAT is characterized by exhibiting high selectivity for arylalkylamines, including indole-ethylamines (e.g. tryptamine, serotonin) and phenyl-ethylamines (e.g. phenylethylamine, norepinephrine, dopamine). The yeast homolog of AANAT can acetylate these same substrates, but with about 1000-fold lower efficiency; in addition, it also acetylates a variety of other amines that are not substrates for the vertebrate AANATs (Ganguly et al., 2001b). The yeast AANAT homolog also acetylates polyamines, which may be the role it plays in this organism (Liu et al., 2005); accordingly, the yeast AANAT homolog has also been termed polyamine N-acetyltransferase. The capacity of vertebrate AANAT to acetylate polyamines has not been established.
The distinctive feature of vertebrate AANAT, the daily rhythm in activity, has not been documented in other groups.
The apparent broader substrate specificity of the yeast AANAT homolog, and the lack of evidence for rhythmic expression is consistent with the view that AANAT homologs function in fungi and bacteria to broadly detoxify intracellular amines (Klein, 2004). Detoxification may have been the advantage conferred when AANAT was acquired by chordates and became associated with the ancestor of the pinealocyte and retinal photoreceptor. AANAT may have made an important contribution to photoreception by acetylating arylalkylamines thereby reducing loss of retinaldehyde via Schiff base formation with arylalkylamines. This would be especially advantageous for dim light detection in the context of a dark environment of the ancestral vertebrate and of limited availability of retinaldehyde.
Vertebrate AANATs are comprised of a catalytic core that binds arylalkylamines and AcCoA and facilitates the transfer of the acetyl group; regulatory regions that contain PKA phosphorylation sites critical for activation and stabilization flank the catalytic core (Fig. 2). Phosphorylation of these sites promotes binding to 14-3-3 proteins, which reduces the Km for the arylalkylamine substrates and also protects the enzyme from proteasomal proteolysis (Ganguly et al., 2001a; Obsil et al., 2001; Ganguly et al., 2005). In addition, a highly conserved lysine in the N-terminal region, may mediate proteasomal proteolysis (Gastel et al., 1998; Klein et al., 1997). The bacterial and fungal AANAT homologs contain some features of the catalytic domain of the vertebrate AANATs (Fig. 2; Ganguly et al., 2001b); however, they lack the flanking regulatory sequences seen in vertebrates.
Figure 2.
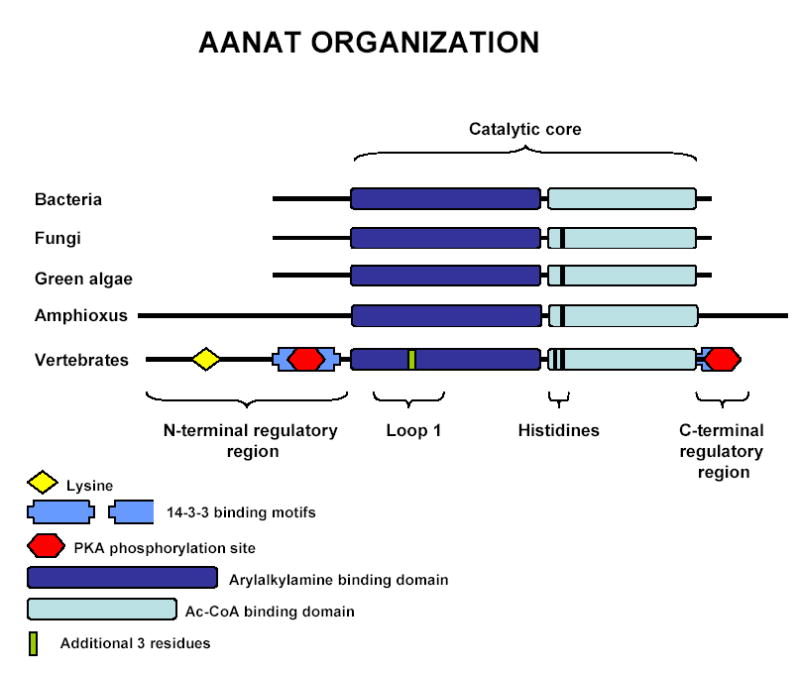
Structural organization of vertebrate AANAT and primitive homologs.
In addition to the changes in the flanking regions during evolution, changes also occurred in the catalytic core (Fig. 2). One is the inclusion of 3 additional amino acids in the Loop 1, which is a flexible element involved in binding of both the arylalkylamine substrate and AcCoA. The additional three residues enhance flexibility and contain a critical proline which forms part of the binding pocket that sandwiches the aromatic group of substrates (Hickman et al., 1999a, 1999b). The absence of these residues from Loop 1 may affect the specificity and/or strength of binding of substrates. A second difference is the addition of a second histidine in a region close to the site of acetyl transfer, thought to promote catalysis by facilitating the removal of protons generated at the active site; mutation of either of these histidines raises the Km significantly and changes the pH dependence (De Angelis et al., 1998; Dyda et al., 2000; Hickman et al., 1999a, 1999b). Vertebrate AANATs contain two histidine groups, one at position 120 in the ovine AANAT sequence and another at position 122, whereas AANAT homologs from fungi have one and those from bacteria have none (Ganguly et al., 2001b; Iyer et al., 2004). The yeast AANAT homolog acetylates arylalkylamines with about 1:1000 the efficiency of vertebrate AANATs which may be partly due to the lack of the second histidine (Ganguly et al., 2001b); little is known regarding the enzyme activity of any of the bacterial homologs.
In the current report we present the results of recent analysis of the available databases, which expands our knowledge of AANAT evolution. Our findings indicate AANAT is present in cephalochordates and in two branches of unicellular algae; in addition, a third AANAT gene is present in fish.
2. Materials and Methods
The GenBank databases (non-redundant and EST) were interrogated with fungal and vertebrate AANATs as query sequences using BLASTP, PsiBLAST and tBLASTn programs (Altschul et al., 1997). Similar searches were performed against raw unassembled sequence from genome sequencing projects (Trace Archives, National Center for Biotechnology Information [NCBI]; http://www.ncbi.nlm.nih.gov/Traces/trace.cgi), and against genomic sequences available at websites dedicated to individual model organisms.
Ancestral AANAT homologs were differentiated from ancestral homologs of other members of the GNAT superfamily by BLAST analysis of a putative AANAT homolog against the GenBank database. Sequences were considered to be AANAT homologs if the query sequence matched best to known AANATs, if the match had an E-value of less than 10−4, and if the significant alignment included regions outside of the AcCoA binding regions, In some cases, apparent exons were assembled into a contiguous open reading frames (ORF). In other cases, a complete ORF was assembled from multiple entries in the Traces Archives.
Multiple sequence alignments were created using ClustalW 1.82, with subsequent manual adjustments.
3. Results and Discussion
3.1. An AANAT homolog in amphioxus, Branchiostoma floridae
An AANAT homolog identified as an EST from an amphioxus embryo cDNA library (GenBank accession no. BW748728) was used to interrogate the B. floridae Trace Archive sequences (NCBI). This identified a set of overlapping traces that were assembled into a continuous ORF (Fig. 3) The trace sequences segregated into two closely related groups; the consensus nucleotide sequences of these two ORFs were 97% identical. These apparently represent two alleles in the amphioxus population.
Figure 3.
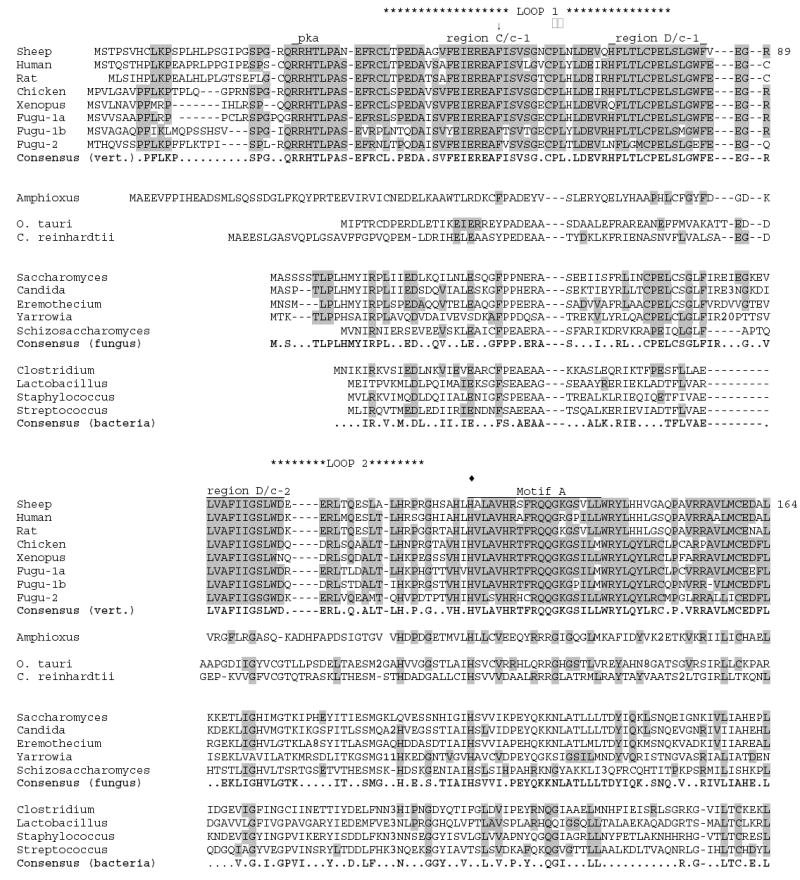

Multiple sequence alignment of putative AANAT homologs from amphioxus, unicellular green alga, fungi, and bacteria compared to vertebrate AANATs. Representative sequences from vertebrates, fungi and bacteria are included. Sequences have been aligned using CLUSTALW, then manually adjusted. Consensus sequences for each phylum are identical to the majority of the sequences shown for that phylum, and are shown in bold. Shaded residues are identical to the vertebrate consensus sequence. Residue numbering is given for sheep AANAT. The sequence included are as follows (numbers are GenBank accession numbers unless otherwise noted): sheep (NP_001009461); human (NP_001079), rat (NP_036950), chicken (NP_990489), Xenopus (allele AANAT-1a1; AAP57668), Fugu-1a, -1b and -2 (assembled from genomic sequence provided at http://genome.jgi-psf.org/Takru4 version 3.0; Aparicio et al., 2002), Amphioxus (assembled from genomic sequence provided in the Trace Archives at the NCBI), O. tauri (predicted peptide product from genomic sequence given in GenBank accession no. CR954223), C. reinhardtii (assembled from genomic sequence provided at http://genome.jgi-psf.org/chlre2), Saccharomyces (NP_010356), Candida (XP_446590), Eremothecium (NP_986794), Yarrowia, (XP_500865), Schizosaccharomyces (CAB57420), Clostridium (AAK76856), Lactobacillus (NP_784135), Staphylococcus (CAI82224), Streptococcus (NP_722057) -- = inserted space; .. = no consensus; bold numbers within sequence = number of residues removed to maintain optimal alignment; □□ = Pro-64 (sheep) part of binding sandwich; □□ = Phe-168 (sheep) part of binding sandwich; ↓ = Phe-56 (sheep) part of binding pocket; ^ = Arg-170 (sheep) part of AcCoA binding; □□ = Tyr-168 (sheep) in catalytic site; □□ = His-122 (sheep) in catalytic site. Regions C/c-1, D/c-1 and D/c-2 are highly conserved in vertebrates as discussed in Klein et al., 1997.
It is important to note that this sequence does not appear to represent a silenced gene or a pseudogene because it is present in an embryonic cDNA library.
Amphioxus is a cephalochordate, a phyla that falls between the most primitive chordate grouping, the urochordates, and vertebrates (Fig. 1). An AANAT homolog has not been found in the genomic sequence of the urochordate, Ciona intestinalis. Likewise extensive searching of recently available genomic sequences has failed to uncover AANAT homologs in any other eukaryote, except fungi and unicellular algae (see below). Thus the amphioxus AANAT homolog reported here is the oldest known AANAT homolog in chordates. This suggests that an identifiable AANAT homolog first appeared in the vertebrate lineage after divergence of the urochordates and - if the HGT hypothesis is correct - was transferred to the chordate line before cephalochordates branched off from the line leading to vertebrates. The absence of introns in the AANAT homologs in amphioxus and fungi is consistent with HGT from bacteria because bacterial AANAT homologs also lack introns. Another hypothetical explanation is that vertebrate AANAT introns were inserted early in chordate evolution after an HGT event and that their absence from cephalochordates reflects a loss which occurred following the divergence of cephalochordates from the vertebrate lineage.
It should be pointed out that the HGT hypothesis is not the only explanation for the biological distribution of AANAT. Another possibility is that ancestral homologs are actually present in many sequenced genomes, but are not recognizable because strong selective pressures may have resulted in rapid divergence. Also, it is possible that AANAT was vertically transferred and repeatedly lost from many lineages, including urochordates (Iyer et al., 2004).
A prominent feature of amphioxus AANAT is that it is longer than vertebrate and fungal homologs at both N- and C-terminal regions (Fig. 2 and 3). These flanking regions, like those in the AANAT homologs in bacteria and fungi, do not contain any of the vertebrate regulatory sequences. This indicates that the regulatory motifs were acquired at a more recent time in chordate evolution. This was advantageous because it enabled rhythmic changes in enzyme activity to occur as a function of posttranslational modifications, i.e. phosphorylation/dephosphorylation which in turn controls the balance between 14-3-3 protection and proteasomal proteolysis. As is true of the yeast AANAT, the cephalochordate AANAT homolog lacks other features of vertebrate AANATs, including the three additional amino acids in Loop 1 and a second histidine in the catalytic core.
3.2. Two AANAT homologs from unicellular green algae
Interrogation of the genomic databases revealed a candidate AANAT homolog in the unicellular green alga, Ostreococcus tauri (predicted peptide product from GenBank entry CR954223; Fig. 3). This nano-alga and is of special interest because it contains the smallest genome currently described among free-living photosynthetic eukaryotic organisms (Derelle et al., 2004). The ORF was found to be intronless, like the amphioxus, fungal and bacterial AANAT homologs. The most similar sequences in GenBank are all vertebrate AANATs, indicating it is a likely AANAT homolog.
Subsequent interrogation of the genomic sequence of another unicellular green alga Chlamydomonas reinhardtii (http://genome.jgi-psf.org/chlre2), using the O. tauri sequence, revealed a related AANAT homolog that contains two introns within the ORF (Fig. 3). These are unrelated to the introns in vertebrate AANATs (Klein et al., 1997).
In contrast to the evidence that AANAT is in algal genomes, AANAT homologs have not been found in the published genomes of higher plants. Accordingly, it is not clear how these homologs were introduced into algae. The algae sequences are similar to those in bacteria, fungi and amphioxus in that they all lack the regulatory flanking sequences, the three vertebrate-linked amino acids in Loop 1, and the second histidine in the catalytic site (Fig. 2). Accordingly, it seems reasonable to suspect that HGT may explain the presence of AANAT in algae, as seems to be the case in fungi and vertebrates (Fig. 1).
3.3. A third AANAT gene in percomorph fishes
As indicated above, only a single AANAT gene is found in mammals, birds and anurans. However, two have been found in teleost fishes: AANAT-1, which is homologous to the non-fish AANATs, is expressed primarily in the fish retina; AANAT-2 is expressed primarily in the fish pineal organ (Bégay et al.,1998; Coon et al., 1999; Benyassi et al., 2000; Falcón et al., 2003; Zilberman-Peled et al., 2004). These two AANAT genes presumably arose from a whole genome duplication early in teleost evolution (Jaillon et al., 2004; Woods et al., 2005). The position of the two introns within the ORF is conserved between the two genes.
Whole genome sequences are available for 4 teleost species. Genome analysis of three of these (two species of pufferfish [Takifugu rubripes and Tetraodon nigroviridis] and of medaka, [Oryzias latipes]) reveal that three AANAT genes are present (Fig. 4). In contrast, a similar analysis of the zebrafish (Danio rerio) genome detected only two genes.
Figure 4.
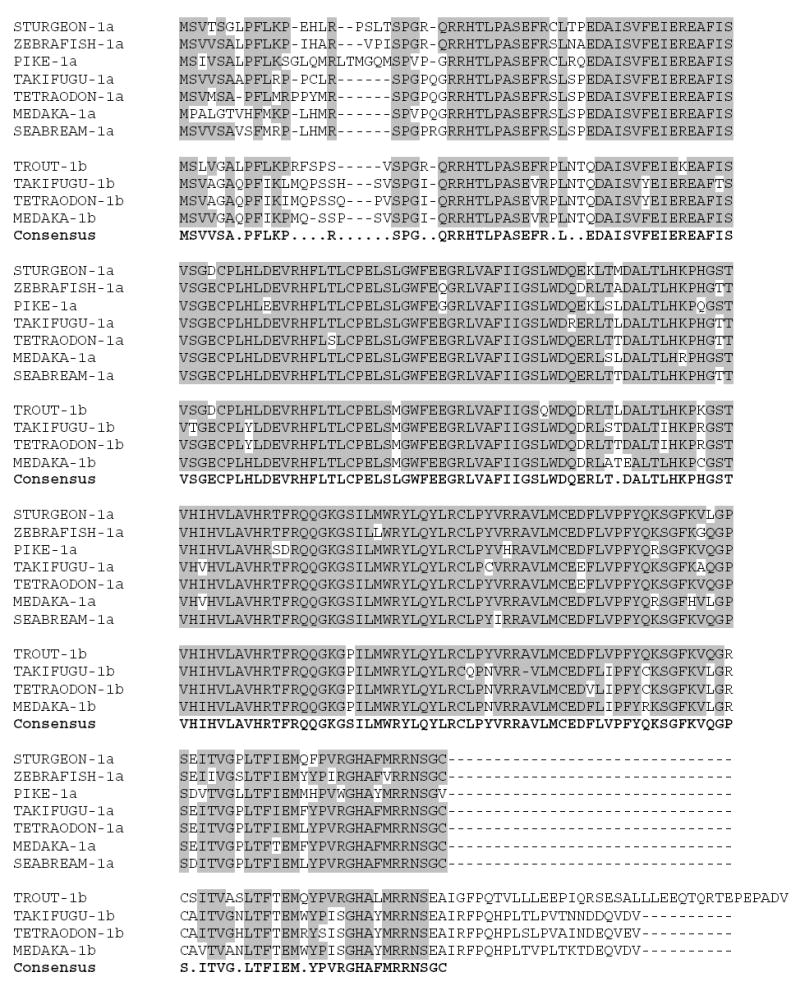
Multiple sequence alignment of fish AANAT-1 peptide sequences in GenBank. Alignment was generated by ClustalW 1.84 followed by minor manual adjustments. The consensus sequence conforms to the majority of included sequences. Shaded residues are identical to the consensus sequence. Peptide sequences were assigned to either the AANAT-1a or AANAT-1b gene as indicated following the label. The sequences included are: STURGEON-1a (Acipenser sturio; GenBank accession no. DQ077733); ZEBRAFISH-1a (Danio rerio; GenBank accession number NM_200704); PIKE-1a (Esox lucius; GenBank accession no. AF034081); TAKIFUGU-1a and -1b (Takifugu rubripes; assembled from genomic sequence provided at http://genome.jgi-psf.org/Takru4 version 3.0; Aparicio et al., 2002); TETRAODON-1a and -1b (Tetraodon nigroviridis; assembled from genomic sequence provided at http://www.genoscope.cns.fr/tetraodon; Jaillon et al., 2004); MEDAKA-1a and -1b (Oryzias latipes;http://dolphi-2005 assembled from genomic sequence provided at http://dolphin.lab.nig.ac.jp/medaka version 200506); SEABREAM-1a (Sparus aurata; GenBank accession no. AY533402); trout-1b (Oncorhynchus mykiss; GenBank accession no. AB007294).
The three genes from pufferfishes and medaka represent two closely related genes corresponding to AANAT-1, termed AANAT-1a and AANAT-1b, and AANAT2. The predicted peptide product of AANAT1a is similar at the C-terminus to all other known vertebrate AANATs and fish AANAT-1s and AANAT-2s. However, the C-terminal regions of AANAT-1b from pufferfish and medaka are 37 residues longer.
The relative substrate specificity of fish AANATs has not been fully established. However, studies of AANAT-2 and AANAT-1a indicate that AANAT-1a acetylates a broad range of substrates with similar avidity, and has a lower Km than AANAT-2, which exhibits relatively higher specificity and a higher Km for indole-ethylamines than AANAT1a (Bégay et al., 1998, Benyassi et al., 2000; Zilberman-Peled et al., 2004).
These characteristics are consistent with AANAT-2 being specialized for melatonin production in the pineal gland. AANAT-1, in contrast, would appear to function to acetylate a range of arylalkylamines in the retina, either for detoxification, biosynthesis of other compounds, or modulation of the amount of an active form of a compound. For example, dopamine is acetylated by AANAT-1a in seabream retina (Zilberman-Peled et al., 2006).
Thus, the existence of multiple AANATs in fish supports the view that the ancestral photoreceptor evolved into the retinal photoreceptor and pinealocyte because of different evolutionary pressures (Klein, 2004), with melatonin synthesis being selected for in the pineal gland and another function in the retinal photoreceptors. Melatonin synthesis requires a high concentration of serotonin, which can interfere with the retinoid cycle during phototransduction in the retina. Conversely, it is thought that AANAT may also act to detoxify arylalkylamines; and, that this was selected for in the retinal photoreceptors. In some mammals, including human, rhesus monkey and cow, the retina has essentially no capacity to make melatonin, but expresses high levels of AANAT activity (Wiechmann, 1993; Rodriquez et al., 1994; Bernard et al., 1995; Coon et al., 1996; Craft et al., 1999; Coon et al., 2002).
The presence of three AANATs in teleosts is likely to be explained by two events. One is the early genome duplication event (Jaillon et al., 2004; Woods et al., 2005), which generated two genes, allowing one to evolve to AANAT-1 and the other to AANAT-2. And, the presence of two AANAT-1s is likely to represent a subsequent gene duplication event. The AANAT-1a and AANAT-1b proteins from the same species are approximately 75–80% identical and 80–85% similar; this degree of difference indicates that the duplicated genes are likely to encode proteins with somewhat different functional characteristics. For comparison, AANAT-1a and AANAT-2 from the same species are 65–70% identical and 70–75% similar.
The evidence that three genes are present in some teleosts indicates that three genes are present in other members of this group, including those that have been used in melatonin research (pike, trout, sea bream and sea bass). In view of this, the results of published studies on AANAT must be revisited.
The forms of AANAT-1 cloned from the pike, Esox lucius (Bégay et al., 1998; GenBank accession no. AF034081), and the seabream, Sparus aurata (Zilberman-Peled et al., 2004; GenBank accession no. AY533402), appear to represent the AANAT-1a gene, since they do not have the C-terminal extension. In comparison, the AANAT-1 cloned from the rainbow trout, Oncorhynchus mykiss, appears to represent the AANAT-1b gene because it possesses an additional 39 amino acids following the C-terminal PKA serine (Mizusawa et al., 1998; GenBank accession no. AB007294). The initial residues in the extension on the trout AANAT-1b exhibit substantial similarity to the extensions on the pufferfish and medaka AANAT-1b proteins (Fig 4) indicating they were derived from a common ancestral gene.
The existence of AANAT-1a and AANAT-1b genes, may explain a number of paradoxes in the literature regarding the expression of AANAT-1. For example, Western blot analysis has revealed a 24 kDa AANAT-1 protein instead of the 28 kDa predicted from the cloned trout AANAT-1 cDNA (Besseau et al., 2006). This might reflect the presence of the shorter AANAT-1a form and the absence AANAT-1b. Another paradox relating to AANAT-1 is that in the seabream 3’- and 5’-RACE analysis of the cloned transcript (now recognized as the shorter AANAT-1a form) predicted a 1.4 kb transcript (Zilberman-Peled et al., 2004). However, Northern blot analysis revealed a single ~ 2.2 kb transcript; this longer transcript could be encoded by the AANAT-1b gene; in this species AANAT-1a may be expressed at only a low level in the retina.
The finding of three AANAT genes is not related to a recent report of two AANAT-1 transcripts in chum salmon (GenBank accession nos. AY356361; AY356362) (Shi et al., 2004). One of which was is expressed in the retina and the other in brain. Comparison of these sequences to the AANAT-1a and AANAT-1b sequences now available indicates that both of the chum salmon sequences are more closely related to AANAT-1b than to AANAT-1a; thus they likely represent an additional gene duplication event in this salmonid. This situation will be become clearer when the full ORF of these transcripts becomes available.
The evidence that both AANAT-1a and AANAT-1b should be present in a wide range of teleosts (at least in salmonids and percomorphs) raises the question of why a second AANAT-1 gene is not present in the zebrafish genome? It is reasonable that the second AANAT-1 gene arose by gene duplication in the main trunk of the teleost linage that occurred after the zebrafish branch had split off (Fig 5). Thus all teleosts after this point would be expected to have two AANAT-1 genes; obviously, loss of one after this point might occur. It is also possible that the gene duplication occurred earlier than the zebrafish branch and that AANAT-1b was subsequently lost from this branch.
Figure 5.
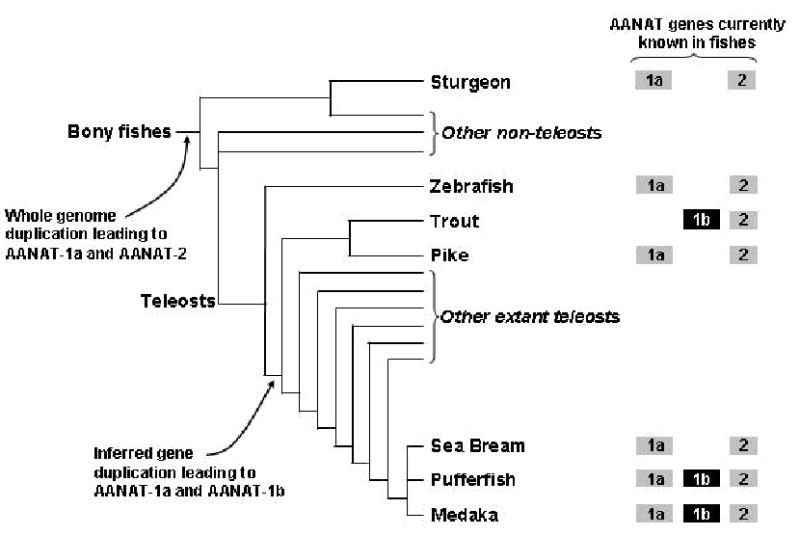
Schematic evolutionary tree for bony fishes showing the approximate relationships between groups of fishes known to contain AANAT genes. It is loosely based on phylogenetic trees shown by the Tree of Life Project (http://tolweb.org) which are in turn based on Nelson (1994). The shaded symbols indicate which AANATs have been identified in each species. Only groups of fish in which an AANAT has been identified have been labeled.
The multiplicity of AANATs in fish may not be a reflection only of genome duplication and gene duplication; it may also reflect two features of reproduction which facilitate rapid genetic changes in response to environmental pressures, i.e. rapid evolution. These features include large progeny size and short generation time, which are common among fishes but not other vertebrates.
4. Final Comment
The results of these studies are of interest for several reasons. First, they highlight the puzzling biological distribution of AANAT. The discovery of AANAT in algae raises the question of whether AANAT was distributed by HGT. The discovery of AANAT in amphioxus is important because it sets the stage for studies directed at determining where and why it is expressed, and whether it is associated with photoreceptors, or is generally distributed, and what are the preferred substrates of this enzyme. The discovery of a third gene in fish and the available data indicate that AANAT reflects the highly adaptive nature of photoreceptors, in that multiple genes evolved and that they exhibit different substrate specificities and are expressed preferentially in either the pineal gland or retina. This is consistent with the view that AANAT serves other functions than melatonin synthesis in the vertebrate retina.
Acknowledgments
This research was supported by funds from the Intramural Research Program of the National Institute of Child Health and Human Development, National Institutes of Health. The medaka genome sequence data has been provided freely by the National Institute of Genetics and the University of Tokyo for use in this publication only.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aparicio S, Chapman J, Stupka E, Putnam N, Chia JM, Dehal P, Christoffels A, Rash S, Hoon S, Smit A, Gelpke MD, Roach J, Oh T, Ho IY, Wong M, Detter C, Verhoef F, Predki P, Tay A, Lucas S, Richardson P, Smith SF, Clark MS, Edwards YJ, Doggett N, Zharkikh A, Tavtigian SV, Pruss D, Barnstead M, Evans C, Baden H, Powell J, Glusman G, Rowen L, Hood L, Tan YH, Elgar G, Hawkins T, Venkatesh B, Rokhsar D, Brenner S. Whole-genome shotgun assembly and analysis of the genome of Fugu rubripes. Science. 2002;297:1301–1310. doi: 10.1126/science.1072104. [DOI] [PubMed] [Google Scholar]
- Arendt J. Chapman and Hall; London: 1995. Melatonin and the Mammalian Pineal Gland. [Google Scholar]
- Bégay V, Falcón J, Cahill GM, Klein DC, Coon SL. Transcripts encoding two melatonin synthesis enzymes in the teleost pineal organ: circadian regulation in pike and zebrafish, but not in trout. Endocrinology. 1998;139:905–912. doi: 10.1210/endo.139.3.5790. [DOI] [PubMed] [Google Scholar]
- Benyassi A, Schwartz C, Coon SL, Klein DC, Falcón J. Melatonin synthesis: arylalkylamine N-acetyltransferases in trout retina and pineal organ are different. Neuroreport. 2000;11:255–258. doi: 10.1097/00001756-200002070-00006. [DOI] [PubMed] [Google Scholar]
- Bernard M, Donohue SJ, Klein DC. Human hydroxyindole-O-methyltransferase in pineal gland, retina and Y79 retinoblastoma cells. Brain Res. 1995;696:37–48. doi: 10.1016/0006-8993(95)00651-6. [DOI] [PubMed] [Google Scholar]
- Besseau L, Benyassi A, Møller M, Coon SL, Weller JL, Boeuf G, Klein DC, Falcón J. Melatonin pathway: breaking the 'high-at-night' rule in trout retina. Exp. Eye Res. 2006;82:620–627. doi: 10.1016/j.exer.2005.08.025. [DOI] [PubMed] [Google Scholar]
- Cahill GM, Grace MS, Besharse JC. Rhythmic regulation of retinal melatonin: metabolic pathways, neurochemical mechanisms, and the ocular circadian clock. Cell Mol. Neurobiol. 1991;11:529–560. doi: 10.1007/BF00734814. [DOI] [PubMed] [Google Scholar]
- Coon SL, Bégay V, Deurloo D, Falcón J, Klein DC. Two arylalkylamine N-acetyltransferase genes mediate melatonin synthesis in fish. J Biol Chem. 1999;274:9076–9082. doi: 10.1074/jbc.274.13.9076. [DOI] [PubMed] [Google Scholar]
- Coon SL, Mazuruk K, Bernard M, Roseboom PH, Klein DC, Rodriguez IR. The human serotonin N-acetyltransferase (EC 2.3.1.87) gene (AANAT): Structure, chromosomal localization, and tissue expression. Genomics. 1996;34:76–84. doi: 10.1006/geno.1996.0243. [DOI] [PubMed] [Google Scholar]
- Coon SL, del Olmo E, Young WS, III, Klein DC. Melatonin synthesis enzymes in Macaca mulatta: focus on arylalkylamine N-acetyltransferase (EC 2.3.1.87) J Clin Endocrinol Metabol. 2002;87:4699–4706. doi: 10.1210/jc.2002-020683. [DOI] [PubMed] [Google Scholar]
- Craft CM, Murage J, Brown B, Zhan-Poe X. Bovine arylalkylamine N-acetyltransferase activity correlated with mRNA expression in pineal and retina. Brain Res Mol Brain Res. 1999;65:44–51. doi: 10.1016/s0169-328x(98)00336-2. [DOI] [PubMed] [Google Scholar]
- De Angelis J, Gastel J, Klein DC, Cole PA. Kinetic analysis of the catalytic mechanism of serotonin N-acetyltransferase (EC 2.3.1.87) J Biol Chem. 1998;273:3045–3050. doi: 10.1074/jbc.273.5.3045. [DOI] [PubMed] [Google Scholar]
- Dyda F, Klein DC, Hickman AB. GCN5-related N-acetyltransferases: a structural overview. Annu Rev Biophys Biomol Struct. 2000;29:81–103. doi: 10.1146/annurev.biophys.29.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falcón J, Gothilf Y, Coon SL, Boeuf G, Klein DC. Genetic, temporal and developmental differences between melatonin rhythm generating systems in the teleost fish pineal organ and retina. J. Neuroendocrinol. 2003;15:378–382. doi: 10.1046/j.1365-2826.2003.00993.x. [DOI] [PubMed] [Google Scholar]
- Ganguly S, Gastel JA, Weller JL, Schwartz C, Jaffe H, Namboodiri MA, Coon SL, Hickman AB, Rollag M, Obsil T, Beauverger P, Ferry G, Boutin JA, Klein DC. Role of a pineal cAMP-operated arylalkylamine N-acetyltransferase/14-3-3-binding switch in melatonin synthesis. Proc Natl Acad Sci U S A. 2001a;98:8083–8088. doi: 10.1073/pnas.141118798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganguly S, Mummaneni P, Steinbach PJ, Klein D C, Coon SL. Characterization of the Saccharomyces cerevisiae homolog of the melatonin rhythm enzyme arylalkylamine N-acetyltransferase (EC 2.3.1.87) J Biol Chem. 2001b;276:47239–47247. doi: 10.1074/jbc.M107222200. [DOI] [PubMed] [Google Scholar]
- Ganguly S, Weller JL, Ho A, Chemineau P, Malpaux B, Klein DC. Melatonin synthesis: 14-3-3-dependent activation and inhibition of arylalkylamine N-acetyltransferase mediated by phosphoserine-205. Proc Natl Acad Sci U S A. 2005;102:1222–1227. doi: 10.1073/pnas.0406871102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gastel JA, Roseboom PH, Rinaldi PA, Weller JL, Klein DC. Melatonin production: proteasomal proteolysis in serotonin N-acetyltransferase regulation. Science. 1998;279:1358–1360. doi: 10.1126/science.279.5355.1358. [DOI] [PubMed] [Google Scholar]
- Hickman AB, Klein DC, Dyda F. Melatonin biosynthesis: the structure of serotonin N-acetyltransferase at 2.5 A resolution suggests a catalytic mechanism. Mol. Cell. 1999a;3:23–32. doi: 10.1016/s1097-2765(00)80171-9. [DOI] [PubMed] [Google Scholar]
- Hickman AB, Namboodiri MA, Klein DC, Dyda F. The structural basis of ordered substrate binding by serotonin N-acetyltransferase: enzyme complex at 1.8 A resolution with a bisubstrate analog. Cell. 1999b;97:361–369. doi: 10.1016/s0092-8674(00)80745-x. [DOI] [PubMed] [Google Scholar]
- Iuvone PM, Tosini G, Pozdeyev N, Haque R, Klein DC, Chaurasia SS. Circadian clocks, clock networks, arylalkylamine N-acetyltransferase, and melatonin in the retina. Prog Retin Eye Res. 2005;24:433–456. doi: 10.1016/j.preteyeres.2005.01.003. [DOI] [PubMed] [Google Scholar]
- Iyer LM, Aravind L, Coon SL, Klein DC, Koonin EV. Evolution of cell-cell signaling in animals: did late horizontal gene transfer from bacteria have a role? Trends Genet. 2004;20:292–299. doi: 10.1016/j.tig.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Jaillon O, Aury JM, Brunet F, Petit JL, Stange-Thomann N, Mauceli E, Bouneau L, Fischer C, Ozouf-Costaz C, Bernot A, Nicaud S, Jaffe D, Fisher S, Lutfalla G, Dossat C, Segurens B, Dasilva C, Salanoubat M, Levy M, Boudet N, Castellano S, Anthouard V, Jubin C, Castelli V, Katinka M, Vacherie B, Biemont C, Skalli Z, Cattolico L, Poulain J, De Berardinis V, Cruaud C, Duprat S, Brottier P, Coutanceau JP, Gouzy J, Parra G, Lardier G, Chapple C, McKernan KJ, McEwan P, Bosak S, Kellis M, Volff JN, Guigo R, Zody MC, Mesirov J, Lindblad-Toh K, Birren B, Nusbaum C, Kahn D, Robinson-Rechavi M, Laudet V, Schachter V, Quetier F, Saurin W, Scarpelli C, Wincker P, Lander ES, Weissenbach J, Roest Crollius H. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Klein DC. The 2004 Aschoff/Pittendrigh lecture: Theory of the origin of the pineal gland--a tale of conflict and resolution. J. Biol. Rhythms. 2004;19:264–279. doi: 10.1177/0748730404267340. [DOI] [PubMed] [Google Scholar]
- Klein DC, Baler R, Roseboom PH, Weller JL, Bernard M, Gastel JA, Zatz M, Iuvone M, Bégay V, Falcón J, Cahill G, Cassone VM, Coon SL. The molecular basis of the pineal melatonin rhythm: Regulation of serotonin N-acetyltransferase. Handbook of Behavioral State Control: Cellular and Molecular Mechanisms. In: Lydic R, Baghdoyan H, editors. CRC Press; Boca Raton: 1998. pp. 45–59. [Google Scholar]
- Klein DC, Coon SL, Roseboom PH, Weller JL, Bernard M, Gastel JA, Zatz M, Iuvone PM, Rodriguez IR, Bégay V, Falcón J, Cahill GM, Cassone VM, Baler R. The melatonin rhythm-generating enzyme: molecular regulation of serotonin N-acetyltransferase in the pineal gland. Recent Prog Horm Res. 1997;52:307–357. [PubMed] [Google Scholar]
- Liu B, Sutton A, Sternglanz R. A yeast polyamine acetyltransferase. J. Biol. Chem. 2005;280:16659–16664. doi: 10.1074/jbc.M414008200. [DOI] [PubMed] [Google Scholar]
- Mizusawa K, Iigo M, Suetake H, Yoshiura Y, Gen K, Kikuchi K, Okano T, Fukuda Y, Aida K. Molecular cloning and characterization of a cDNA encoding the retinal arylalkylamine N-acetyltransferase of the rainbow trout, Oncorhynchus mykiss. Zool Sci. 1998;15:345–351. doi: 10.2108/zsj.15.345. [DOI] [PubMed] [Google Scholar]
- Nelson JS. 3rd Ed. John Wiley and Sons; New York, N.Y.: 1994. Fishes of the World. [Google Scholar]
- Obsil T, Ghirlando R, Klein DC, Ganguly S, Dyda F. Crystal structure of the 14-3-3zeta:serotonin N-acetyltransferase complex. A role for scaffolding in enzyme regulation. Cell. 2001;105:257–267. doi: 10.1016/s0092-8674(01)00316-6. [DOI] [PubMed] [Google Scholar]
- Rodriguez IR, Mazuruk K, Schoen TJ, Chader GJ. Structural analysis of the human hydroxyindole-O-methyltransferase gene. Presence of two distinct promoters. J. Biol. Chem. 1994;269:31969–31977. [PubMed] [Google Scholar]
- Shi Q, Ando H, Coon SL, Sato S, Ban M, Urano A. Embryonic and post-embryonic expression of arylalkylamine N-acetyltransferase and melatonin receptor genes in the eye and brain of chum salmon (Oncorhynchus keta) Gen Comp Endocrinol. 2004;136:311–321. doi: 10.1016/j.ygcen.2004.01.004. [DOI] [PubMed] [Google Scholar]
- Tosini G. Melatonin circadian rhythm in the retina of mammals. Chronobiol. Int. 2000;17:599–612. doi: 10.1081/cbi-100101067. [DOI] [PubMed] [Google Scholar]
- Tosini G, Menaker M. Circadian rhythms in cultured mammalian retina. Science. 1996;272:419–421. doi: 10.1126/science.272.5260.419. [DOI] [PubMed] [Google Scholar]
- Vetting MW, de Carvalho LPS, Yu M, Hegde SS, Magnet S, Roderick SL, Blanchard JS. Structure and functions of the GNAT superfamily of acetyltransferases. Arch. Biochem. Biophys. 2005;433:212–226. doi: 10.1016/j.abb.2004.09.003. [DOI] [PubMed] [Google Scholar]
- Wiechmann AF. Localization of hydroxyindole-O-methyltransferase in the retina: a re-evaluation. Exp Eye Res. 1993;57:379–380. doi: 10.1006/exer.1993.1137. [DOI] [PubMed] [Google Scholar]
- Woods IG, Wilson C, Friedlander B, Chang P, Reyes DK, Nix R, Kelly PD, Chu F, Postlethwait JH, Talbot WS. The zebrafish gene map defines ancestral vertebrate chromosomes. Genome Res. 2005;15:1307–1314. doi: 10.1101/gr.4134305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zilberman-Peled B, Benhar I, Coon SL, Ron B, Gothilf Y. Duality of serotonin-N-acetyltransferase in the gilthead seabream (Sparus aurata): molecular cloning and characterization of recombinant enzymes. Gen Comp Endocrinol. 2004;138:139–147. doi: 10.1016/j.ygcen.2004.05.007. [DOI] [PubMed] [Google Scholar]
- Zilberman-Peled B, Ron B, Gross A, Finberg JPM, Gothilf Y. Brain Res. 2006. A possible new role for fish retinal serotonin-N-acetyltransferase-1 (AANAT1): Dopamine metabolism. In press. [DOI] [PubMed] [Google Scholar]


