Figure 3.
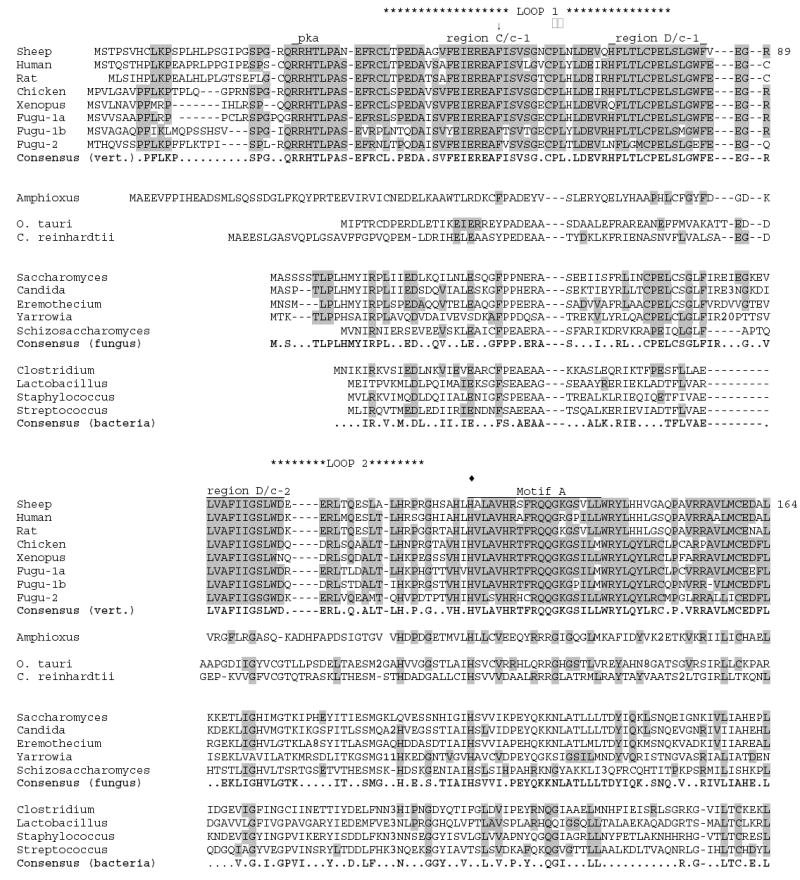

Multiple sequence alignment of putative AANAT homologs from amphioxus, unicellular green alga, fungi, and bacteria compared to vertebrate AANATs. Representative sequences from vertebrates, fungi and bacteria are included. Sequences have been aligned using CLUSTALW, then manually adjusted. Consensus sequences for each phylum are identical to the majority of the sequences shown for that phylum, and are shown in bold. Shaded residues are identical to the vertebrate consensus sequence. Residue numbering is given for sheep AANAT. The sequence included are as follows (numbers are GenBank accession numbers unless otherwise noted): sheep (NP_001009461); human (NP_001079), rat (NP_036950), chicken (NP_990489), Xenopus (allele AANAT-1a1; AAP57668), Fugu-1a, -1b and -2 (assembled from genomic sequence provided at http://genome.jgi-psf.org/Takru4 version 3.0; Aparicio et al., 2002), Amphioxus (assembled from genomic sequence provided in the Trace Archives at the NCBI), O. tauri (predicted peptide product from genomic sequence given in GenBank accession no. CR954223), C. reinhardtii (assembled from genomic sequence provided at http://genome.jgi-psf.org/chlre2), Saccharomyces (NP_010356), Candida (XP_446590), Eremothecium (NP_986794), Yarrowia, (XP_500865), Schizosaccharomyces (CAB57420), Clostridium (AAK76856), Lactobacillus (NP_784135), Staphylococcus (CAI82224), Streptococcus (NP_722057) -- = inserted space; .. = no consensus; bold numbers within sequence = number of residues removed to maintain optimal alignment; □□ = Pro-64 (sheep) part of binding sandwich; □□ = Phe-168 (sheep) part of binding sandwich; ↓ = Phe-56 (sheep) part of binding pocket; ^ = Arg-170 (sheep) part of AcCoA binding; □□ = Tyr-168 (sheep) in catalytic site; □□ = His-122 (sheep) in catalytic site. Regions C/c-1, D/c-1 and D/c-2 are highly conserved in vertebrates as discussed in Klein et al., 1997.
