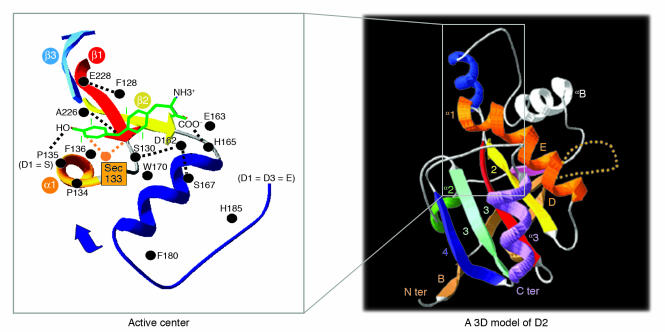
Figure 2. Deiodinases.
While the deiodinases have not yet been crystallized, protein modeling indicates that they share a common general structure composed of a single aminoterminal-anchoring segment, a short hinge region, and a thioredoxin fold–containing globular domain (9). A 3D model of the D2 globular domain is shown on the right. Letters and numbers shown indicate different β sheets and α-helices as previously reported (9). The orange dotted loop indicates the D2-specific segment that mediates interaction with the E3-ubiquitin ligase WSB-1 (28). The inset illustrates the active center, which contains the rare amino acid selenocysteine (Sec), which is critical for nucleophilic attack during the deiodination reaction. The residues that putatively interact with the T4 molecule (green) are also shown. Position 135, which in D2 and D3 is occupied by proline, is critical for enzyme kinetics. D2 and D3 have high affinity for their substrates and are not sensitive to inhibition by PTU. Replacement with serine, which is naturally found in D1, turns both D2 and D3 into low affinity and PTU-sensitive enzymes (9). C ter, C terminus; N ter, N terminus. Figure modified with permission from the Journal of Biological Chemistry (9) and Nature Cell Biology (28).