Abstract
Increasing multidrug resistance in Enterococcus faecalis, a nosocomial opportunist and common cause of bacterial endocarditis, emphasizes the need for alternative therapeutic approaches such as immunotherapy or immunoprophylaxis. In an earlier study, we demonstrated the presence of antibodies in E. faecalis endocarditis patient sera to recombinant forms of 9 E. faecalis cell wall–anchored proteins; of these, we have now characterized an in vivo–expressed locus of 3 genes and an associated sortase gene (encoding sortase C; SrtC). Here, using mutation analyses and complementation, we demonstrated that both the ebp (encoding endocarditis and biofilm-associated pili) operon and srtC are important for biofilm production of E. faecalis strain OG1RF. In addition, immunogold electron microscopy using antisera against EbpA–EbpC proteins as well as patient serum demonstrated that E. faecalis produces pleomorphic surface pili. Assembly of pili and their cell wall attachment appeared to occur via a mechanism of cross-linking of the Ebp proteins by the designated SrtC. Importantly, a nonpiliated, allelic replacement mutant was significantly attenuated in an endocarditis model. These biologically important surface pili, which are antigenic in humans during endocarditis and encoded by a ubiquitous E. faecalis operon, may be a useful immunotarget for studies aimed at prevention and/or treatment of this pathogen.
Introduction
Enterococci, generally considered as normal bowel commensals, are also recognized as opportunistic pathogens (1) and rank among the top 3 causes of nosocomial bloodstream, surgical site, and urinary tract infections (2). Among enterococci, the most clinically abundant species, Enterococcus faecalis, accounts for approximately 5%–8% of hospital-associated bacteremia and approximately 5%–20% of all cases of endocarditis (3). Endocarditis is an infection of the heart’s valves or inner lining that leads to valvular destruction and death without effective antibiotic therapy. The acquisition of resistance to multiple antibiotics has made enterococcal endocarditis a life-threatening clinical challenge, and this highlights the need for alternatives to current antibiotic strategies (4). In this regard, alternative approaches that might be considered include immunoprophylaxis or immunotherapy targeting proteins that are expressed in vivo and that are important for virulence.
The first step in the infection process of extracellular pathogens like E. faecalis is generally considered to be attachment to and colonization of host tissue surfaces. Evidence from other gram-positive pathogens suggests that proteins from the microbial surface component recognizing adhesive matrix molecules (MSCRAMM) family may serve as potential antigenic candidates for the development of novel immunotherapies (5). We have recently identified 17 proteins with cell wall–anchoring motifs having MSCRAMM-like structural features (6) from the E. faecalis V583 genome (7). These proteins consist of 1 or more regions of 150- to 500-aa segments containing Ig-like fold(s) characteristic of the Ig family of Staphylococcus aureus MSCRAMMs (6, 8). The demonstration of the presence of antibodies in sera from patients with E. faecalis endocarditis to at least some of these proteins indicates that they are indeed expressed in vivo during infection; most E. faecalis patient sera showed particularly high titers against 3 of these proteins, namely EbpA, EbpB, and EbpC (endocarditis and biofilm-associated pili; previously referred to as EF1091, EF1092, and EF1093, respectively; ref. 6).
Besides MSCRAMM-mediated colonization, another factor that is predicted to be important in E. faecalis infection is the ability of strains to form biofilm (9–11). Our recent systematic study that analyzed biofilm formation by a large number of E. faecalis isolates found that endocarditis isolates produced biofilm significantly more often and also to a greater degree than nonendocarditis isolates (12). To date, at least 7 genes or gene clusters have been implicated in biofilm formation by E. faecalis (12–17).
Among gram-negative bacteria (e.g., uropathogenic E. coli and Neisseria spp.), it has long been known that some accomplish adhesion through the use of fibrous protein organelle(s) present on the bacterial surface, named pili or fimbriae (18), but only recently has limited information on gram-positive surface organelles such as pili been reported (19–22). However, the physiological role of these gram-positive pili in infectious processes such as endocarditis has yet to be demonstrated. Based on recent publications of pilus-like structures on Corynebacterium diphtheriae and Streptococcus spp., it appears that these gram-positive pili are formed by ordered cross-linking of multiple different classes of precursor proteins that are tethered by designated sortases (19–22). Sortases were previously recognized as transamidases that covalently anchored proteins with a C-terminal, LPXTG-like motif to the peptidoglycan of gram-positive bacteria (23). Using a bioinformatics approach, Ton-That et al. (19, 24) predicted pilin-associated motifs in other gram-positive bacteria, including a single protein, EbpC, of E. faecalis. Among E. faecalis electron microscopy studies, a single study — in 1981 — reported fimbriae-like structures on the surface of E. faecalis strain JH2 (25) using negative staining.
Since our previous study found high titers of antibodies against 3 proteins (encoded by contiguous genes) in sera obtained from patients with E. faecalis infections (6), in this study, we characterized ef1091–ef1093 at the molecular level. We introduced mutations into each of the ef1091–ef1094 loci and analyzed the importance of these genes for virulence. Our results demonstrated that the products of the cotranscribed ef1091–ef1093 genes form surface pili that are dependent on sortase (also referred to as EF1094) and that these structures play a role in biofilm formation as well as in endocarditis.
Results
A disruption mutant of ebpA is defective in biofilm formation.
In an attempt to assign a phenotype to ef1091, ef1092, and ef1093 loci (6), shown to be antigenic during serious E. faecalis infections such as endocarditis, we first constructed an insertion disruption mutation in the ef1091 locus of E. faecalis strain OG1RF (a medium-biofilm producer; biofilm density [OD570], 1–2) and, using a polystyrene microtiter plate assay, found that an ef1091 disruption mutant of OG1RF showed significant reduction (P < 0.0001) in biofilm formation (Supplemental Figure 1) to below our cutoff (12) for a non–biofilm producer. We subsequently designated this gene ebpA based on the functions identified herein. The growth pattern of the ebpA disruption mutant (TX5421; see Supplemental Table 1; supplemental material available online with this article; doi:10.1172/JCI29021DS1) was essentially the same as wild-type OG1RF in brain/heart infusion broth (BHI; routine growth medium for enterococci) as well as in tryptic soy broth with glucose (TSBG; medium used for biofilm assay). Since ebpA is part of a predicted operon in the sequenced genome of E. faecalis V583, the observed biofilm-negative phenotype of the ebpA disruption mutant may be due to interruption of the ebpA gene and/or polar effects on the expression of gene(s) downstream of ebpA. Hence, we analyzed this region from E. faecalis OG1RF.
Organization of the ebp locus of E. faecalis OG1RF.
Analysis of the 8,694-bp nucleotide region of OG1RF spanning ebpA–ebpC and srtC plus 500 bp on each side predicted the presence of 4 complete open reading frames (ORFs) similar to the published sequence of E. faecalis V583 (Figure 1A). Just 3 bp downstream of the 3,432-bp ebpA gene is the 1,431-bp ebpB gene. The 1,884-bp ebpC gene, encoding a 627-aa protein, overlaps the ebpB gene by 4 bp. Downstream of ebpC, separated by 93 bp, is an 855-bp predicted sortase gene, sortase C (srtC). Comparison of ebpA–ebpC and srtC sequences from strain OG1RF with the sequences from strain V583 revealed 99.0%–99.4% identity at the DNA level and 98.7%–99.4% identity at the protein level. Within the 8,694-bp region there were 65 nucleotide differences, of which many were silent. All 3 Ebp proteins contain features characteristic of MSCRAMMs (26) and in silico structural predictions as well as far-UV circular dichroism measurements suggest that these proteins are likely to adopt Ig-like fold(s) such as those found in the characterized MSCRAMMs of S. aureus (6, 8).
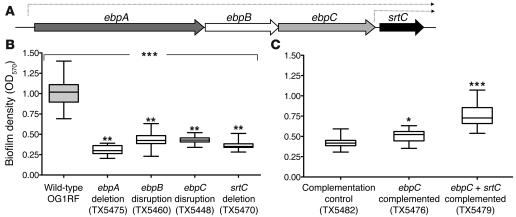
Figure 1. Biofilm formation and the ebpA–srtC operon.
(A) Illustration of the ebpA–srtC operon of E. faecalis OG1RF. (B) Comparison of biofilm production of wild-type OG1RF and ebpA deletion (TX5475), ebpB disruption (TX5460), ebpC disruption (TX5448), and srtC deletion (TX5470) mutants. (C) Biofilm formation after complementing the ebpC disruption mutant with ebpC (TX5476) and ebpC plus srtC (TX5479) cloned under control of the nisin-inducible promoter of pMSP3545. The ebp disruption mutant electroporated with vector only (TX5482) served as a control. Median and interquartile range values are shown. Values are from at least 3 independent experiments, each performed in quadruplicate. *P = 0.0079, **P < 0.001, ***P < 0.0001; Kruskal-Wallis test (ANOVA) for comparing WT with mutants and Mann-Whitney U test for comparing complemented strains with vector-only control.
The genes ebpA to srtC are transcribed as a polycistronic operon.
Besides our prediction of ebpA–ebpC cotranscription due to short or overlapping intergenic regions, we were unable to identify transcriptional terminator–like sequences in the region from ebpA to srtC. RT-PCR analysis using intergenic bridging primer sets amplified transcripts for the regions between ebpA and ebpB, ebpB and ebpC, and ebpC and srtC (Supplemental Figure 2, lanes 4, 7, and 10), indicating that these genes are linked and cotranscribed as a polycistronic mRNA. Thus, these data indicate that the ebp locus represents a 4-gene operon.
RT-PCR analysis of the ebpA disruption mutant showed highly reduced transcripts of downstream ebpB and ebpC genes; however, transcripts for srtC were only moderately reduced compared with wild-type OG1RF, which suggests that there is an independent srtC transcript in addition to the srtC present on an ebpA–ebpC transcript (Supplemental Figure 3A). The incomplete polar effect observed in the ebpA disruption mutant was likely due to the presence of weak promoter activity in the truncated ebpA gene or in the vector. RNA analysis of a subsequently made ebpB disruption mutant (TX5460; see Supplemental Table 1) showed complete absence of transcripts of ebpC, but only a moderate reduction in transcripts for srtC compared with wild-type OG1RF (seen also with the ebpC disruption mutant), supporting this conclusion (Supplemental Figure 3, B–D). Consistent with this result, transcriptional start site and promoter analysis identified candidate promoter sequences upstream of ebpA and srtC start codons (Supplemental Figure 4). Of note, no effect on ebpA transcript levels was found in ebpB, ebpC, or srtC mutants, indicating that these genes were not involved in autoregulation under our testing conditions.
The role of the ebp locus in biofilm formation of OG1RF.
Polar disruption mutants of ebpB as well as ebpC (TX5448; see Supplemental Table 1) were found to be stable, while insertion in srtC exhibited some instability. To overcome this problem, we subsequently deleted the complete srtC gene from OG1RF (TX5470; see Supplemental Methods and Supplemental Table 1). We also constructed a polar ebpA deletion mutant of OG1RF (TX5475; see Supplemental Table 1) by substituting a kanamycin resistance cassette. All ebp and srtC mutant strains and the parental strain OG1RF demonstrated equivalent growth kinetics as well as similar gross morphology and chain length in both BHI and TSBG (data not shown). Examination of the biofilm-forming capacity of these mutants revealed that the ebpA deletion mutant, the ebpB disruption mutant, the ebpC disruption mutant, and the srtC deletion mutant were markedly attenuated in their ability to form a biofilm (P < 0.001), similar to the ebpA disruption mutant (Figure 1B). Taken together, these results indicate that biofilm formation by E. faecalis OG1RF is affected by the ebpA–srtC operon.
Since biofilm formation is a multistep procedure (27), with the first step being attachment to biotic and abiotic surfaces, the wild-type and mutant strains were scored for differences in primary attachment using phase-contrast microscopy. Significant differences were seen in initial attachment step of biofilm formation of ebpA, ebpB, ebpC, and srtC mutants (Supplemental Figure 5).
An intact ebp locus is required for optimal biofilm formation of OG1RF.
To confirm that the ebpA–ebpC mutations were responsible for the reduced biofilm formation and to exclude a possible independent effect due only to SrtC, we introduced a wild-type copy of ebpC or of ebpC plus srtC (cloned into shuttle vector pMSP3545 with a nisin-inducible promoter; ref. 28) into each of the ebpA–ebpC and srtC mutants. As vector-only controls, mutants were electroporated with pMSP3545. Introduction of ebpC into the ebpC disruption mutant (TX5476; see Supplemental Table 1) showed a small increase in biofilm formation, whereas complementation with ebpC plus srtC (TX5479; see Supplemental Table 1) substantially restored biofilm. The ebpC disruption mutant electroporated with pMSP3545 (TX5482; see Supplemental Table 1) retained its reduced biofilm phenotype (Figure 1C). Complementation of the srtC deletion mutant with ebpC plus srtC completely restored biofilm formation (Supplemental Figure 6A). On the other hand, introduction of these constructs into both the ebpA deletion mutant and the ebpB disruption mutant did not restore biofilm formation (Supplemental Figure 6, B and C). Taken together, these results indicate that biofilm formation by E. faecalis OG1RF requires expression of both the ebpA–ebpC operon and the srtC gene.
Ebp proteins form a SrtC-dependent, heteromeric, high–molecular weight complex.
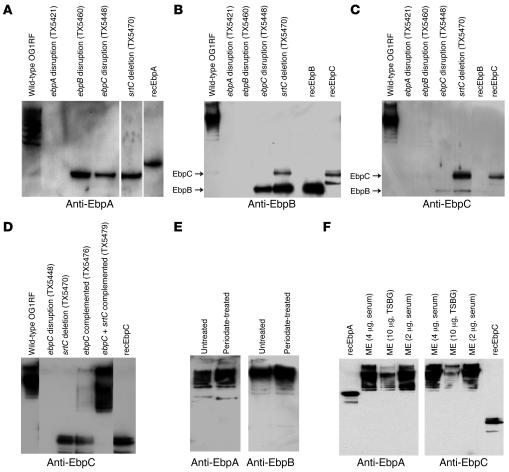
Muramidase-derived surface protein extracts of OG1RF analyzed using gradient polyacrylamide gels and probed with anti-EbpA antiserum detected a ladder of high–molecular weight (HMW) bands (EbpAHMW) with masses greater than 200 kDa, in addition to a faint immunoreactive band of approximately 140 kDa, slightly larger than the calculated EbpA mass of 120.4 kDa (Figure 2A). No bands were seen with the ebpA disruption mutant (Figure 2A). Probing surface extracts of the ebpB and ebpC disruption mutants as well as the srtC deletion mutant with anti-EbpA serum detected a single band (~140 kDa) likely representing monomeric EbpA, indicating that multiple proteins are required for formation of EbpAHMW (Figure 2A). Anti-EbpB and anti-EbpC sera as well as recombinant EbpB and EbpC affinity-purified antibodies also revealed HMW species with surface protein extracts of OG1RF (Figure 2, B and C). Probing of extracts from the 4 mutants with either anti-EbpB or anti-EbpC antibodies also showed absence of HMW species and reaction with monomeric bands representing upstream genes, as anticipated; the complemented strain, with partial to largely restored biofilm, also showed partial restoration of HMW bands (Figure 2D). We observed strong cross-reactivity between anti-EbpB–specific Igs and EbpC, and some cross-reactivity between anti-EbpC–specific Igs and EbpB, likely due to the homology between these proteins (Supplemental Figure 7). Since we observed similar HMW laddering with Epa-associated polysaccharides of E. faecalis OG1RF (29), we also analyzed these muramidase-derived surface preparations for the presence of carbohydrates by periodate treatment, and results indicated that this HMW ladder was not caused by carbohydrates (Figure 2E).
Figure 2. Ebp proteins form an SrtC-dependent HMW ladder.
(A–D) Immunoblots of mutanolysin extracts of wild-type E. faecalis OG1RF; ebpA disruption (TX5421), ebpB disruption (TX5460), ebpC disruption (TX5448), and srtC deletion (TX5470) mutants; and ebpC disruption mutants complemented with ebpC (TX5476) or ebpC plus srtC (TX5479) probed with anti-Ebp antibodies. rec-, recombinant. (E) OG1RF mutanolysin extracts, either left untreated or treated with periodate, were probed with anti-Ebp sera. (F) Immunoblots of varying amounts of mutanolysin extracts (ME) of OG1RF grown in either TSBG or 40% horse serum and probed with anti-Ebp antibodies. The affinity-purified anti-Ebp Igs or anti-Ebp sera used in each Western blot are indicated. The srtC deletion mutant sample in A and the ebpB disruption mutant sample in B are from different gels.
When grown in biofilm medium (TSBG), HMW Ebp bands were produced at levels detectable with Western blot even after ×100 concentration of surface extracts by lyophilization. Growth in routine medium (BHI) produced much lower levels of HMW Ebp bands. However, semiquantitative RT-PCR failed to detect significant differences among ebp transcript levels. Among other conditions tested, one that enhanced levels of HMW Ebp bands in Western blots was growth in serum (Figure 2F), a biological cue with in vivo relevance. RT-PCR after growth in serum showed elevated levels of the independent srtC transcript without an apparent increase in ebp mRNA levels (S.R. Nallapareddy et al., unpublished observations).
We next sought to determine whether other sortases play a role in the formation of HMW Ebp bands. Analysis of the preliminary genomic sequence of OG1RF generated by our collaboration with the Human Genome Sequencing Center at Baylor College of Medicine (G. Weinstock, B.E. Murray, D.A. Garsin, et al., unpublished genome) and hybridization studies indicate that only 2 of the 3 sortases of strain V583 are present in strain OG1RF. Disruption of the gene encoding the other sortase (SrtA; also known as EF3056) of OG1RF had no effect on the production of HMW Ebp bands (data not shown). Taken together, our results indicate that the associated SrtC polymerizes monomers of EbpA, EbpB, and EbpC to generate HMW Ebp bands.
Identification of conserved motifs in Ebp proteins.
The spectrum of HMW bands and the reported pilin motif–like sequence and E-box of C. diphtheriae pilin proteins in EbpC (19, 24) led us to look for similar motifs in EbpA and EbpB proteins. Both motifs were found in both proteins. In addition, EbpA and EbpC were each found to have 2 regions with features common to E-box motifs (Supplemental Figure 8). The lysine residue of the pilin motif and the glutamic acid of the E-box, essential for polymerization of C. diphtheriae pilus (19, 24), were both conserved. No additional ORFs with these motifs were found either in the published sequence of E. faecalis V583 nor in the preliminary genome sequence of E. faecalis OG1RF. Based on this information, we predicted that the ebpA–srtC operon codes for pilus-like surface structures.
The ebp locus mediates production of surface-attached pili, detectable by immunogold staining.
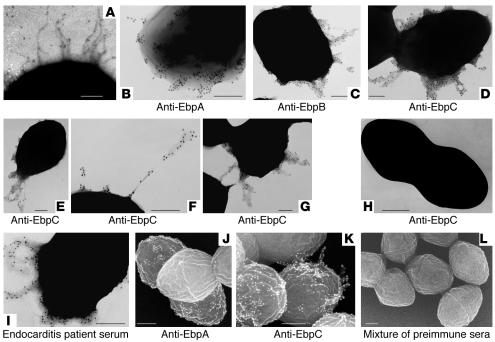
High-resolution transmission electron microscopy (TEM) of negatively stained E. faecalis OG1RF cells grown in BHI showed pilus-like surface structures in less than 1% of cells (Figure 3A). When TSBG-grown cells were negatively stained, a dense, tangled mass of material was seen surrounding the surface of the cells, and this material obscured the demarcation of pili. However, anti-EbpA, -EbpB, and -EbpC sera and affinity-purified anti-EbpB and -EbpC Igs, together with 12 nm gold-labeled secondary antibody, reacted with appendages extending from the surface of OG1RF grown in TSBG (Figure 3). These data suggest that all 3 Ebp proteins are components of the E. faecalis pili. Anti-Ebp–stained pili were seen in 10%–20% cells in different growth phases of TSBG-grown OG1RF cells, with the greatest number seen at the entry into the stationary phase. However, anti-Ebp staining of BHI-grown OG1RF cells detected pili in rare cells (0% to <2%). Preimmune rabbit sera (with OG1RF) and ebpA, ebpC, and srtC mutants served as controls and showed no pili (ebpC disruption mutant is shown in Figure 3H).
Figure 3. Ebp proteins form SrtC-dependent pili.
(A) Pili on the surface of E. faecalis. Strain OG1RF grown in BHI was negatively stained with uranyl acetate and viewed by TEM. (B–G) Immunogold labeling and TEM of OG1RF pili stained with anti-Ebp antibodies. (H) Immunogold labeling and TEM of the ebpC disruption mutant stained with anti-EbpC antibodies. (I) Immunogold labeling and TEM of OG1RF stained with endocarditis patient serum. (J–L) Immunogold labeling and SEM of OG1RF pili stained with anti-Ebp antibodies. The anti-Ebp antisera used in each micrograph is indicated. The mixture of the 3 preimmune sera is shown as SEM control. Scale bars: 200 nm.
It was also observed that in some cells, Ebp-associated pili were peritrichous (Figure 3D), while in others they either appeared as a tangled mass of multiple thin filaments or seemed localized to a particular part of the cell, such as a pole (Figure 3E); occasionally, these pili were long (Figure 3F). Pili often appeared to be associated with more than one cell (Figure 3G). Of interest, serum obtained from a patient with E. faecalis endocarditis also stained the pili (Figure 3I).
We also investigated OG1RF and ebp mutant strains with low-voltage scanning electron microscopy (SEM). The results from TEM negative staining studies were corroborated by 3D SEM views (Figure 3), and in both, EbpA staining was most intense at the base of the pili and only rarely appeared along the shaft (Figure 3, B and J). This was in contrast to staining with affinity-purified anti-EbpB and -EbpC Igs, which showed immunogold staining throughout the pilus (Figure 3, C–G and K). Taken together, our results suggest that E. faecalis pili are formed by ordered cross-linking of 3 different precursor proteins, each with conserved motif sequences that may be recognized by a designated sortase, SrtC.
Ebp pili contribute to E.
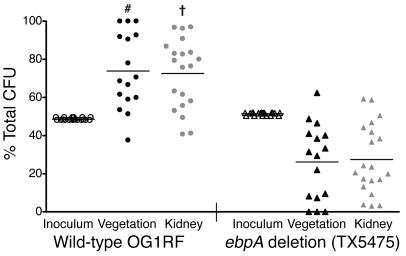
faecalis endocarditis. Since the infective endocarditis vegetation is a biofilm process on heart valves, we tested the nonpiliated ebpA deletion mutant in the rat endovascular infection model. This model has been widely used to assess bacterial adherence to damaged heart valves. After estimating the optimum inoculum, the ebpA deletion mutant and wild-type OG1RF were injected as an equal mixture, as described in previous studies (30, 31). Figure 4 shows the proportions of wild-type OG1RF to ebpA deletion mutant CFUs in the inoculum (t = 0; OG1RF comprised slightly less than 50%), and at t = 24 hours in vegetation and kidneys. Mean ratios of wild-type OG1RF to the ebpA deletion mutant among bacteria recovered from vegetation and kidneys were 0.739:0.261 and 0.726:0.274, respectively (P = 0.0069 and P = 0.0093 for the percentages of OG1RF in the inoculum versus the percentages of OG1RF in vegetation and kidneys, respectively), demonstrating outnumbering of the mutant by wild-type. The mean virulence indices (32) of the mutant relative to OG1RF were 0.06 and 0.27 in vegetation and kidneys, respectively, indicating that Ebp pili play an important role in this endovascular infection.
Figure 4. Attenuation of a nonpiliated ebpA deletion mutant in an endocarditis model.
Percentages of viable wild-type OG1RF and the ebpA deletion mutant (TX5475) recovered from vegetation, kidneys, and initial inocula from 20 rats are shown; 4 rats had sterile vegetations. Horizontal lines indicate means. #P = 0.0069, †P = 0.0093 versus inoculum; χ2 test.
The ebp locus is ubiquitous among E.
faecalis strains. Using specific probes for ebpA, ebpB, and ebpC, we tested colony lysates of 408 well-characterized E. faecalis isolates from 4 continents and found that all 408 E. faecalis isolates were positive under high-stringency conditions. The presence of genes encoding immunogenic proteins in 100% of our worldwide collection spanning 30 years suggests that they are part of the core genome and may be important for survival or proliferation of E. faecalis. Using the publicly available E. faecium genome sequence generated in collaboration with the Human Genome Sequencing Center at Baylor College of Medicine (http://www.hgsc.bcm.tmc.edu/projects/microbial/Efaecium), we also found that a similar locus with identical gene organization was present in that species as well.
Discussion
With increasing antibiotic resistance, interest has increased in the use of bacterial factors that are important for an organism’s ability to colonize, infect, and eventually cause disease as immunotherapeutic targets (5). Among these factors, MSCRAMMs, which are implicated in initiating infections including endocarditis (33), have drawn considerable attention lately because of their widespread nature in traditional and opportunistic pathogens (5, 26). The isolation and characterization of MSCRAMMs from E. faecalis has had limited success, because this organism does not readily adhere to ECM proteins under laboratory growth conditions (34, 35), unlike its more aggressive relatives staphylococci and streptococci. Therefore, we recently used a bioinformatics approach and identified several proteins that predict MSCRAMM-like structures (6). Based on reactivity with sera from E. faecalis–infected patients, we inferred that some of these predicted proteins are indeed expressed by E. faecalis during infection. Furthermore, antibodies against 3 of these proteins were found at significantly higher levels in the sera of most infected patients (6), thus making them attractive candidates for the present study.
Here we report that the widespread ebp genes encoding the above-mentioned in vivo–expressed and antigenic proteins are in a polycistronic operon and that this operon contains an additional gene coding for sortase. We also demonstrated a role for the ebp locus in the production of biofilm using E. faecalis OG1RF. We chose OG1RF for characterization of this locus for several reasons. First, it represents the medium-biofilm category (biofilm density [OD570], 1–2), which was shown by our previous study systematically categorizing E. faecalis biofilm formation of 163 diverse isolates to be the most common category, displayed by 56% of isolates (12). Second, this strain is well characterized (36) and amenable to genomic manipulations (29, 37) and, most importantly, animal models including endocarditis have been established for this strain (38, 39). Mutations within the ebpA–srtC gene cluster showed markedly reduced biofilm formation. These data, combined with complementation studies, indicate that both the ebpA–ebpC operon and the srtC gene are required for biofilm formation of OG1RF. Importantly, the failure of the ebpB disruption mutant (defective in EbpB and EbpC and partially defective in SrtC) with ebpC-srtC to restore biofilm indicates that reduced biofilm formation by ebp mutants is not due to a secondary, independent effect of SrtC. Furthermore, the differences seen in initial attachment step of biofilm formation indicate that these genes play a role in the early cell-surface interactions of the multistep biofilm formation process.
Biofilm formation is considered important in various infections including endocarditis and is a particular concern because of the potential to further protect already drug-resistant organisms from antibiotics and opsonophagocytosis and to allow increased horizontal gene transfer (10, 40). Previously published E. faecalis studies have shown that disruptions of esp (encoding enterococcal surface protein) of some strains, the fsr 2-component system, gelE (encoding gelatinase), epa gene cluster (encoding the polysaccharide Epa), atn (encoding an autolysin), or bopD (sugar-binding transcriptional regulator) resulted in a significant reduction in biofilm density (12–16). A recent study that identified the traits involved in the high-biofilm–producing strain E99 by transposon mutagenesis found a gene cluster named bee with an organization similar to that of Ebp (17). However, this gene cluster was found to be rare in E. faecalis and appeared to be localized to a conjugative plasmid. Similarly, esp, gelE, and fsr loci are also restricted to a subpopulation of E. faecalis. Furthermore, gene association studies using clinical isolates have shown that neither esp (12) nor fsr/gelE (41) is required for E. faecalis biofilm formation. This observed variation in the presence or absence of multiple biofilm-related genes may partially account for the variable capacity of biofilm formation (i.e., strong producers to nonproducers) seen within this species.
To confirm the predicted Ebp localization to the E. faecalis cell surface, we analyzed cell surface preparations of OG1RF and its isogenic ebp and srt mutants. Western blot analysis using anti-Ebp antibodies revealed multiple periodate-resistant HMW bands with all 3 anti-Ebp antibodies; in contrast, only a single band was seen in cell surface preparations with the E. faecalis collagen-binding MSCRAMM, Ace (37). HMW Ebp bands were visible in wild-type OG1RF and in constructs expressing all 4 products of the ebp locus, but not in the mutants, indicating that both srtC and the ebpA–ebpC operon are required for these HMW Ebp bands. The observation of a similar laddering pattern linked to pili in other gram-positive bacteria (19–22) and the identification of motifs conserved in pilus-forming proteins in all 3 Ebps suggested that Ebp proteins might also form pilus-like structures.
Immunogold electron microscopy studies showed that E. faecalis OG1RF produced peritrichous pili, and our results suggest that all 3 Ebp proteins are components of the E. faecalis surface pili. The observed association of pili with more than one cell also suggests there could be an additional cell-cell interaction function of Ebp in the multistep biofilm formation process. To eliminate false positives (commonly seen during negative staining), immunostaining was carried out in eppendorf tubes followed by multiple gentle washes prior to addition of cells to the EM grids. Both SEM 3D views and TEM negative staining studies showed the most intense staining of EbpA at the pilus base. This is in contrast to anti-EbpB and -EbpC Igs, which showed immunogold staining throughout the pili. Since anti-EbpB Igs are strongly cross-reactive with EbpC, no definitive conclusions can be drawn about the location of EbpB. Based on our demonstration of the uniform distribution of EbpC along the pilus shaft using anti-EbpC and the previously observed strong similarity of EbpC with the major subunits of other gram-positive pili-forming proteins (19, 24), it is likely that EbpC is a major component of the shafts of E. faecalis pili. Taken together, these observations suggest that similar to the recently described gram-positive pilus formation mechanism of C. diphtheriae (19), E. faecalis pili appear to be formed by ordered cross-linking of 3 different Ebp proteins, each with conserved motif sequences that are recognized by a designated sortase, SrtC. This mechanism is different from the proposed polymerization of glycosylated Fap1 fimbriae of S. parasanguis (18) and other oral pathogens.
It is interesting to note that anti-Ebp–stained pili were seen in less than 20% of OG1RF cells when grown in TSBG and in many fewer cells (0% to <2%) after growth in BHI. Since there is no striking difference in the ebp transcript levels of cells grown in these media, the different production of pili in these microenvironments may be controlled at the posttranscriptional level or by factors yet to be elucidated. We speculate that the presence of antigenic pili on just a portion of cells may be advantageous to the organism, permitting transmigration or persistence at infection sites without stimulating strong inflammatory responses.
Infective endocarditis due to multidrug-resistant E. faecalis is the most serious and clinically challenging of all E. faecalis–related infections. Since the endocarditic vegetation is considered a biofilm process on heart valves, we tested a nonpiliated ebpA deletion mutant in the endocarditis model. Rats challenged with a mixture of equal numbers of wild-type E. faecalis OG1RF and its isogenic ebpA deletion mutant showed vegetation, with the percentages of nonpiliated mutants recovered from vegetation and kidneys 24 hours after inoculation being significantly lower than the percentages of wild type, indicating that Ebp pili play an important role in this endovascular infection, in parallel to our in vitro biofilm results. The precise mechanism of action in causing endocarditis has yet to be elucidated. Interestingly, while Ebp pili are produced in a fraction of cells (at least in vitro), they contribute significantly to the pathogenicity of E. faecalis, which underscores the possibility of using Ebp proteins as immunotargets.
In conclusion, the identification of immunogenic, pleomorphic surface pili that were unnoticed during years of E. faecalis virulence studies and their association with biofilm and endocarditis adds a new perspective to the present molecular view of E. faecalis pathogenesis. The ubiquitous nature of this surface appendage–encoding gene cluster in E. faecalis isolates and the finding of a similar locus in a multidrug-resistant endocarditis-derived E. faecium strain have important implications for possible development of an immunoprophylactic or therapeutic approach to clinically challenging multidrug-resistant enterococci.
Methods
Bacterial strains.
All the constructs of wild-type and mutant E. faecalis OG1RF, E. coli, and plasmids used in this study are listed in Supplemental Table 1. All bacterial constructs were given TX numbers, and plasmids from these constructs were assigned pTEX numbers. A brief description on 408 diverse E. faecalis strains that were used to establish the widespread nature of the ebp locus is provided in Supplemental Methods.
Mutagenesis and complementation.
Target genes in E. faecalis OG1RF were disrupted using the suicide vector pTEX4577 (39) containing an internal fragment of the genes of interest. The primers used to amplify internal fragments of ebpA, ebpB, and ebpC (designated ef1091, ef1092, and ef1093, respectively, in the E. faecalis V583 genome database; ref. 7) are listed in Supplemental Table 2. E. faecalis OG1RF was transformed by electroporation, and transformants were selected on Todd-Hewitt agar plates containing kanamycin (2,000 μg/ml) (39). Correct allelic disruption was confirmed by PCR analysis followed by pulsed field gel electrophoresis and Southern blots (42, 43). Details on the construction of ebpA and srtC deletion mutants of OG1RF are provided in Supplemental Methods.
To complement the ebpC disruption mutant in trans, a 1,986-bp fragment containing the ebpC ORF plus its ribosome-binding site (amplified using primers Ef1093comF and Ef1093comR; see Supplemental Table 2) as well as a 3,037-bp fragment containing the ebpC and srtC ORFs (amplified using primers Ef1093comF and Ef1094comR; see Supplemental Table 2) were cloned under the control of the nisin promoter of the shuttle vector, pMSP3545 (28). The resulting constructs were transformed into ebp and srtC mutants by electroporation (44). The expression of ebpC and srtC genes in the ebpC strains complemented with ebpC and ebpC plus srtC after nisin induction (28) was determined by RT-PCR.
Biofilm assay and primary attachment.
A biofilm density detection assay was carried out as described previously (12). Briefly, bacteria were grown overnight, diluted 1:100 in 200 μl TSBG containing 0.25% glucose, and inoculated into polystyrene microtiter plates (BD Biosciences — Falcon). After 24 hours of static incubation at 37°C, attached cells from plates were processed, fixed with Bouin’s fixative for 30 minutes, stained with 1% crystal violet for 30 minutes, and rinsed with distilled water. Crystal violet was solubilized in ethanol/acetone (80:20, vol/vol), and OD570 was determined. Each assay was performed in quadruplicate at least 3 times. For complemented and vector-only control constructs, nisin was added to a final concentration of 25 ng/ml during 24 hours’ static incubation. For the primary attachment assay, cultures that were diluted overnight (OD600 adjusted to 0.1) were added to polystyrene dishes (BD Biosciences — Falcon) and incubated for 2 hours as described previously (13). Bacteria in 25 different fields were counted using phase-contrast microscopy.
RT-PCR.
Total RNA for use in RT-PCR reactions was isolated from TSBG-grown E. faecalis cultures using RNeasy Mini Kit (Qiagen). Total RNA (20–40 μg) was treated 3 times with 20 U RQ1 DNase (Promega) for 30 minutes at 37°C, and DNase was removed using the RNeasy Mini Kit (Qiagen) and purification protocol. Gel electrophoresis for assessing RNA quality was performed as previously described (45). Total RNA (1–50 ng) was reverse transcribed with specific primers (Supplemental Table 2) using the SuperScript One-Step RT-PCR with Platinum Taq kit (Invitrogen) according to the manufacturer’s instructions. As an internal control, a 528-bp fragment of gdh (encoding glyceraldehyde-3-phosphate dehydrogenase) was amplified using the gdhF and gdhR primers (Supplemental Table 2). Reactions without RT were used as controls to detect DNA contamination in total RNA.
Recombinant proteins and antibodies.
Construction of recombinant plasmids as well as expression and purification of recombinant proteins used in this study were as described previously (6). Rabbit polyclonal sera specific for recombinant EbpA, EbpB, and EbpC (previously referred to as EF1091, EF1092, and EF1093, respectively; ref. 6) proteins were generated by Bethyl Laboratories. Specific anti-Ebp Igs were prepared using respective recombinant Ebp proteins as immunoadsorbents and uniflow 4 agarose (Sterogene Bioseparations Inc.) per the manufacturer’s protocol.
Cell wall–associated protein extraction and Western blotting.
Surface protein extracts from E. faecalis strains were prepared using mutanolysin as described previously (46) with minor modifications. In brief, the bacterial pellets from TSBG-grown cells (8 h; 37°C; starting inoculum, ~0.01 OD600) or 40% horse serum–grown cells (10 h; 37°C; starting inoculum, ~0.02 OD600) were washed with 0.02 M Tris-HCl, pH 7.0, and 0.01 M MgSO4 buffer and resuspended in one-tenth the volume of the same buffer containing 100 μM PMSF. Following the addition of mutanolysin to a final concentration of 10 U/1 OD600 of cells, tubes were incubated at 37°C for 1 hour in a rotating shaker. After centrifugation at 12,000 g for 10 minutes, the supernatants were concentrated by lyophilization, and aliquots were stored at 70°C. Equal concentrations of mutanolysin extracts were electrophoresed in 4%–12% NuPAGE Novex Bis-Tris Gels (Invitrogen) under reducing conditions in MOPS buffer and transferred to a PVDF membrane according to the manufacturer’s protocol. Membranes were then probed with anti-EbpA, anti-EbpB, and anti-EbpC polyclonal antisera and/or preimmune sera (primary antibody) followed by HRP-conjugated goat anti-rabbit IgG antibodies (secondary antibody) and developed using ECL detection reagents (Amersham Biosciences).
Electron microscopy and immunogold labeling.
Negative staining experiments were carried out as described previously for C. diphtheriae (24), with some modifications. Briefly, bacterial strains grown overnight in BHI or TSBG were reinoculated to a final OD600 of 0.1 and were grown to different phases, then gently washed in 0.1 M NaCl and stained with 0.1% uranyl acetate.
For immunogold labeling, 1-ml volumes of appropriately grown bacteria were pelleted, washed gently with PBS using a wide-bore tip, and resuspended in PBS containing 5% goat serum. Cells were incubated with a 1:100 dilution of rabbit serum raised against EbpA (1:500 dilution for anti-EbpB and anti-EbpC sera) for 2 hours. The cells were washed 3 times with PBS with 5% goat serum and then incubated with 1:100 dilution of goat anti-rabbit IgG conjugated to 12-nm-diameter colloidal gold particles (Jackson ImmunoResearch Laboratories Inc.) for 1 hour. After 3 subsequent washes with PBS, bacteria were resuspended gently in 2% glutaraldehyde in 0.1 M sodium cacodylate buffer as a fixative.
For TEM examination, the immunogold-labeled cells in fixative were floated on carbon-coated grids for 15 minutes and stained with 0.1% uranyl acetate for 30 seconds. After being washed in saline, the grids were examined with a JEOL 1200 TEM at magnifications of ×30,000–×100,000. For SEM examination, the immunogold-labeled cells in fixative were placed on 5-mm × 10-mm glass supports covered by poly- l-lysine (Sigma-Aldrich). The samples were then washed in 0.15 M cacodylate buffer and postfixed for 90–120 minutes in 1% OsO4 in 0.15 M cacodylate buffer. Samples were then rinsed in cacodylate buffer and dehydrated in an ascending ethanol series (50%, 70%, 80%, 95%, and 100% twice) before critical-point drying with CO2. Specimens were mounted on adhesive carbon films and then coated with approximately 1 nm of platinum using an Ion Tech argon ion beam coater. Samples were examined in a Hitachi S-4700 Field Emission SEM.
Experimental endocarditis in rats.
As a control for differences that may be seen in individual rats (e.g., differences in defense mechanisms or differences in catheter positioning and resulting heart valve damage) that may influence endocarditis results, we used mixing experiments with equal ODs of wild-type and mutant bacteria; this method has previously been shown to be sensitive for detecting virulence differences (30–32). Vegetation was produced in male Sprague-Dawley rats (~250 g) by inserting a polyethylene catheter (Intramedic PE 10; Braintree Scientific Inc.) into the right carotid artery, across the aortic valve, and into the left ventricle. Surgical procedures were carried out on anesthetized animals. Twenty minutes later, the inoculum was injected intraventricularly through the catheter, the catheter was heat sealed and imbedded in subcutaneous tissue, and the skin was closed. Wild-type OG1RF and the ebpA deletion mutant, each representing approximately 50% of bacterial cells, were thoroughly mixed prior to administration into rats. Colony counts of the inoculum were determined by serial dilution and plating on BHI agar with and without kanamycin (2,000 μg/ml), followed by 24 hours of incubation at 37°C. For 6 animals, the CFU counts of wild-type OG1RF and the ebpA deletion mutant in the inoculum were 3.25 × 107 and 3.30 × 107, respectively; thus, the percentage of wild-type OG1RF in the total inoculum was 49.6%. For 9 animals, the CFU counts of wild-type OG1RF and the ebpA deletion mutant were 3.82 × 108 and 4.12 × 108, respectively; thus, the percentage of wild-type OG1RF in the total inoculum was 48.1 %. For 5 animals, the CFU counts of wild-type OG1RF and the ebpA deletion mutant were 4.35 × 108 and 4.65 × 108, respectively; thus, the percentage of wild-type OG1RF in the total inoculum was 48.3%. At 24 hours after inoculation, animals were sacrificed, and aortic vegetations were excised with aseptic technique, weighed, homogenized in 1 ml normal saline, and serially diluted for colony counts. Recovery of samples from kidneys followed a similar procedure, except these were homogenized in 5 ml saline. Colonies were counted after 24 hours’ incubation, and log10 CFU/g of vegetation or kidneys were compared. As previously described for other organisms in mixed infections (32, 47), the mean virulence index of the mutant relative to wild-type was calculated using the following equation:
where GM-CFU indicates geometric mean CFU in the initial inoculum (t = 0) or in vegetation and kidneys (t = 24 hours).
All experimental protocols were approved prior to the beginning of the study by the Animal Welfare Committee of the University of Texas Health Science Center at Houston.
Statistics.
OD570 medians from biofilm assays of wild-type and mutant strains were compared by Kruskal-Wallis test (ANOVA) with Dunn’s post-hoc modification for multiple comparisons. Medians of complemented strains were compared with appropriate vector-only control strains by Mann-Whitney test. Tests were performed using GraphPad Prism version 4.0 for Windows. The percentages of OG1RF in the inoculum versus the percentages of OG1RF in vegetation and kidneys were compared using results for individual rats by the χ2 test.
Supplementary Material
Acknowledgments
This work was supported by NIH grants R37 AI47923, from the Division of Microbiology and Infectious Diseases of NIAID (to B.E. Murray), and AI20624 (to M. Hook). We thank Karen Jacques-Palaz and Shahreen Chowdhury for their technical assistance and Agathe Bourgogne for her discussions.
Footnotes
Nonstandard abbreviations used: BHI, brain/heart infusion broth; Ebp, endocarditis and biofilm-associated pili; HMW, high–molecular weight; MSCRAMM, microbial surface component recognizing adhesive matrix molecules; ORF, open reading frame; SEM, scanning electron microscope, scanning electron microscopy; SrtC, sortase C; TEM, transmission electron microscope, transmission electron microscopy; TSBG, tryptic soy broth with glucose.
Conflict of interest: The authors have declared that no conflict of interest exists.
Citation for this article: J. Clin. Invest. 116:2799–2807 (2006). doi:10.1172/JCI29021
Jouko Sillanpää’s present address is: Division of Infectious Diseases, Department of Internal Medicine, University of Texas Medical School at Houston, Houston, Texas, USA.
Sreedhar R. Nallapareddy and Kavindra V. Singh contributed equally to this work.
Stanley L. Erlandsen is deceased. This paper is dedicated to the memory of this coauthor, without whom this work would not have been possible.
See the related Commentary beginning on page 2582.
References
- 1.Murray B.E., Weinstock G.M. Enterococci: new aspects of an old organism. Proc. Assoc. Am. Physicians. 1999;111:328–334. doi: 10.1046/j.1525-1381.1999.99241.x. [DOI] [PubMed] [Google Scholar]
- 2.Richards M.J., Edwards J.R., Culver D.H., Gaynes R.P. Nosocomial infections in combined medical-surgical intensive care units in the United States. Infect. Control Hosp. Epidemiol. 2000;21:510–515. doi: 10.1086/501795. [DOI] [PubMed] [Google Scholar]
- 3.Megran D.W. Enterococcal endocarditis. Clin. Infect. Dis. 1992;15:63–71. doi: 10.1093/clinids/15.1.63. [DOI] [PubMed] [Google Scholar]
- 4.Koch S., Hufnagel M., Theilacker C., Huebner J. Enterococcal infections: host response, therapeutic, and prophylactic possibilities. Vaccine. 2004;22:822–830. doi: 10.1016/j.vaccine.2003.11.027. [DOI] [PubMed] [Google Scholar]
- 5.Rivas J.M., Speziale P., Patti J.M., Hook M. MSCRAMM-targeted vaccines and immunotherapy for staphylococcal infection. Curr. Opin. Drug Discov. Devel. 2004;7:223–227. [PubMed] [Google Scholar]
- 6.Sillanpaa J., Xu Y., Nallapareddy S.R., Murray B.E., Hook M. A family of putative MSCRAMMs from Enterococcus faecalis. . Microbiology. 2004;150:2069–2078. doi: 10.1099/mic.0.27074-0. [DOI] [PubMed] [Google Scholar]
- 7.Paulsen I.T., et al. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. . Science. 2003;299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- 8.Deivanayagam C.C., et al. A novel variant of the immunoglobulin fold in surface adhesins of Staphylococcus aureus: crystal structure of the fibrinogen-binding MSCRAMM, clumping factor A. . EMBO J. 2002;21:6660–6672. doi: 10.1093/emboj/cdf619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Toole G., Kaplan H.B., Kolter R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- 10.Carniol K., Gilmore M.S. Signal transduction, quorum-sensing, and extracellular protease activity in Enterococcus faecalis biofilm formation. . J. Bacteriol. 2004;186:8161–8163. doi: 10.1128/JB.186.24.8161-8163.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Donlan R.M., Costerton J.W. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 2002;15:167–193. doi: 10.1128/CMR.15.2.167-193.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mohamed J.A., Huang W., Nallapareddy S.R., Teng F., Murray B.E. Influence of origin of isolates, especially endocarditis isolates, and various genes on biofilm formation by Enterococcus faecalis. . Infect. Immun. 2004;72:3658–3663. doi: 10.1128/IAI.72.6.3658-3663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toledo-Arana A., et al. The enterococcal surface protein, Esp, is involved in Enterococcus faecalis biofilm formation. . Appl. Environ. Microbiol. 2001;67:4538–4545. doi: 10.1128/AEM.67.10.4538-4545.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hancock L.E., Perego M. The Enterococcus faecalis fsr two-component system controls biofilm development through production of gelatinase. . J. Bacteriol. 2004;186:5629–5639. doi: 10.1128/JB.186.17.5629-5639.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hufnagel M., Koch S., Creti R., Baldassarri L., Huebner J. A putative sugar-binding transcriptional regulator in a novel gene locus in Enterococcus faecalis contributes to production of biofilm and prolonged bacteremia in mice. . J. Infect. Dis. 2004;189:420–430. doi: 10.1086/381150. [DOI] [PubMed] [Google Scholar]
- 16.Kristich C.J., Li Y.H., Cvitkovitch D.G., Dunny G.M. Esp-independent biofilm formation by Enterococcus faecalis. . J. Bacteriol. 2004;186:154–163. doi: 10.1128/JB.186.1.154-163.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tendolkar P.M., Baghdayan A.S., Shankar N. Putative surface proteins encoded within a novel transferable locus confer a high-biofilm phenotype to Enterococcus faecalis. . J. Bacteriol. 2006;188:2063–2072. doi: 10.1128/JB.188.6.2063-2072.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu H., Fives-Taylor P.M. Molecular strategies for fimbrial expression and assembly. Crit. Rev. Oral Biol. Med. 2001;12:101–115. doi: 10.1177/10454411010120020101. [DOI] [PubMed] [Google Scholar]
- 19.Ton-That H., Marraffini L.A., Schneewind O. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. . Mol. Microbiol. 2004;53:251–261. doi: 10.1111/j.1365-2958.2004.04117.x. [DOI] [PubMed] [Google Scholar]
- 20.Mora M., et al. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15641–15646. doi: 10.1073/pnas.0507808102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lauer P., et al. Genome analysis reveals pili in Group B Streptococcus. Science. 2005;309:105. doi: 10.1126/science.1111563. [DOI] [PubMed] [Google Scholar]
- 22.Barocchi M.A., et al. A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl. Acad. Sci. U. S. A. 2006;103:2857–2862. doi: 10.1073/pnas.0511017103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mazmanian S.K., Liu G., Ton-That H., Schneewind O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. . Science. 1999;285:760–763. doi: 10.1126/science.285.5428.760. [DOI] [PubMed] [Google Scholar]
- 24.Ton-That H., Schneewind O. Assembly of pili on the surface of Corynebacterium diphtheriae. . Mol. Microbiol. 2003;50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- 25.Handley P.S., Jacob A.E. Some structural and physiological properties of fimbriae of Streptococcus faecalis. . J. Gen. Microbiol. 1981;127:289–293. doi: 10.1099/00221287-127-2-289. [DOI] [PubMed] [Google Scholar]
- 26.Foster T.J., Hook M. Surface protein adhesins of Staphylococcus aureus. . Trends Microbiol. 1998;6:484–488. doi: 10.1016/s0966-842x(98)01400-0. [DOI] [PubMed] [Google Scholar]
- 27.Dunne W.M. Bacterial adhesion: seen any good biofilms lately? Clin. Microbiol. Rev. 2002;15:155–166. doi: 10.1128/CMR.15.2.155-166.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bryan E.M., Bae T., Kleerebezem M., Dunny G.M. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid. 2000;44:183–190. doi: 10.1006/plas.2000.1484. [DOI] [PubMed] [Google Scholar]
- 29.Xu Y., Murray B.E., Weinstock G.M. A cluster of genes involved in polysaccharide biosynthesis from Enterococcus faecalis OG1RF. . Infect. Immun. 1998;66:4313–4323. doi: 10.1128/iai.66.9.4313-4323.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Winberg J., et al. The PapG-adhesin at the tip of P-fimbriae provides Escherichia coli with a competitive edge in experimental bladder infections of cynomolgus monkeys. . J. Exp. Med. 1995;182:1695–1702. doi: 10.1084/jem.182.6.1695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hienz S.A., Schennings T., Heimdahl A., Flock J.I. Collagen binding of Staphylococcus aureus is a virulence factor in experimental endocarditis. . J. Infect. Dis. 1996;174:83–88. doi: 10.1093/infdis/174.1.83. [DOI] [PubMed] [Google Scholar]
- 32.Nannini E.C., Teng F., Singh K.V., Murray B.E. Decreased virulence of a gls24 mutant of Enterococcus faecalis OG1RF in an experimental endocarditis model. . Infect. Immun. 2005;73:7772–7774. doi: 10.1128/IAI.73.11.7772-7774.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moreillon P., Que Y.A., Bayer A.S. Pathogenesis of streptococcal and staphylococcal endocarditis. Infect. Dis. Clin. North Am. 2002;16:297–318. doi: 10.1016/s0891-5520(01)00009-5. [DOI] [PubMed] [Google Scholar]
- 34. Gilmore, M.S., Coburn, P.S., Nallapareddy, S.R., and Murray, B.E. 2002. Enterococcal virulence. In The enterococci: pathogenesis, molecular biology, antibiotic resistance and infection control. M.S. Gilmore, et al., editors. American Society for Microbiology. Washington, DC, USA. 301–354. [Google Scholar]
- 35.Xiao J., Hook M., Weinstock G.M., Murray B.E. Conditional adherence of Enterococcus faecalis to extracellular matrix proteins. . FEMS Immunol. Med. Microbiol. 1998;21:287–295. doi: 10.1111/j.1574-695X.1998.tb01176.x. [DOI] [PubMed] [Google Scholar]
- 36.Murray B.E., et al. Generation of restriction map of Enterococcus faecalis OG1 and investigation of growth requirements and regions encoding biosynthetic function. . J. Bacteriol. 1993;175:5216–5223. doi: 10.1128/jb.175.16.5216-5223.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nallapareddy S.R., Qin X., Weinstock G.M., Hook M., Murray B.E. Enterococcus faecalis adhesin, ace, mediates attachment to extracellular matrix proteins collagen type IV and laminin as well as collagen type I. . Infect. Immun. 2000;68:5218–5224. doi: 10.1128/iai.68.9.5218-5224.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Singh K.V., Nallapareddy S.R., Nannini E.C., Murray B.E. Fsr-independent production of protease(s) may explain the lack of attenuation of an Enterococcus faecalis fsr mutant versus a gelE-sprE mutant in induction of endocarditis. . Infect. Immun. 2005;73:4888–4894. doi: 10.1128/IAI.73.8.4888-4894.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Singh K.V., Qin X., Weinstock G.M., Murray B.E. Generation and testing of mutants of Enterococcus faecalis in a mouse peritonitis model. . J. Infect. Dis. 1998;178:1416–1420. doi: 10.1086/314453. [DOI] [PubMed] [Google Scholar]
- 40.Parsek M.R., Singh P.K. Bacterial biofilms: an emerging link to disease pathogenesis. Annu. Rev. Microbiol. 2003;57:677–701. doi: 10.1146/annurev.micro.57.030502.090720. [DOI] [PubMed] [Google Scholar]
- 41.Mohamed J.A., Murray B.E. Lack of correlation of gelatinase production and biofilm formation in a large collection of Enterococcus faecalis isolates. . J. Clin. Microbiol. 2005;43:5405–5407. doi: 10.1128/JCM.43.10.5405-5407.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qin X., et al. Targeted mutagenesis of enterococcal genes. Methods Cell Sci. 1998;20:21–33. [Google Scholar]
- 43.Murray B.E., Singh K.V., Heath J.D., Sharma B.R., Weinstock G.M. Comparison of genomic DNAs of different enterococcal isolates using restriction endonucleases with infrequent recognition sites. J. Clin. Microbiol. 1990;28:2059–2063. doi: 10.1128/jcm.28.9.2059-2063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li X., Weinstock G.M., Murray B.E. Generation of auxotrophic mutants of Enterococcus faecalis. . J. Bacteriol. 1995;177:6866–6873. doi: 10.1128/jb.177.23.6866-6873.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sambrook, J., Fritsch, E.F., and Maniatis, J. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press. Cold Spring Harbor, New York, USA. 1659 pp. [Google Scholar]
- 46.Nallapareddy S.R., Singh K.V., Duh R.W., Weinstock G.M., Murray B.E. Diversity of ace, a gene encoding a microbial surface component recognizing adhesive matrix molecules, from different strains of Enterococcus faecalis and evidence for production of ace during human infections. . Infect. Immun. 2000;68:5210–5217. doi: 10.1128/iai.68.9.5210-5217.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beuzon C.R., Holden D.W. Use of mixed infections with Salmonella strains to study virulence genes and their interactions in vivo. Microbes Infect. 2001;3:1345–1352. doi: 10.1016/s1286-4579(01)01496-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.