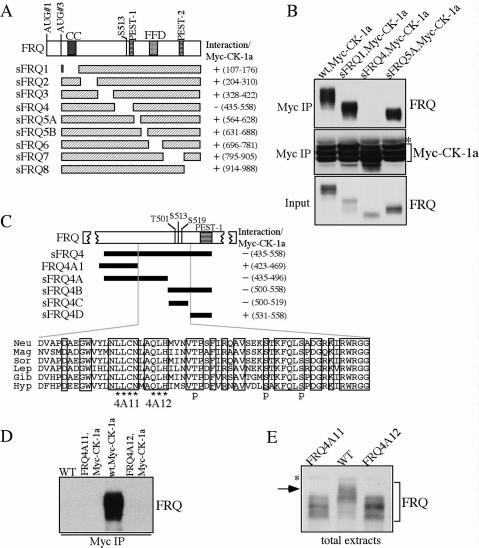
Figure 1.
Disruption of the FRQ/CK-1a results in the hypophosphorylation of FRQ. (A) Graphic diagrams showing the domain structure of FRQ and different sFRQ internal deletion mutants. (CC) Coiled-coil domain; (FFD) FRQ–FRH interaction domain; (S513) a previously identified FRQ phosphorylation site. FRQ amino acids deleted in each mutant are indicated. (B) Immunoprecipitation assay showing that sFRQ4 failed to interact with Myc-CK-1a. The asterisk indicates the IgG signal. (C) Graphic diagrams showing FRQ/CK-1a interaction domain (FCD) and the internal deletions of FRQ in various mutants. The black bars indicate the regions deleted in the different mutants. FRQ amino acids deleted in each mutant are indicated. Except for FRQ4A1, which expresses full-length FRQ protein with the indicated deletion, other mutants only express sFRQ. The amino acid alignment of the FCD domains of different FRQ homologs is shown at bottom and the identical residues among different homologs are boxed. (Neu) Neurospora crassra; (Mag) Magnaporthe grisea; (Sor) Sordaria fimicola; (Lep) Leptosphaeria australiensis; (Gib) Gibberella zeae; (Hyp) Hypocrea spinulosa. In the 4A11 and 4A12 mutants (express full-length FRQ), the amino acids indicated by the asterisks were mutated to alanines. (p) The three previously identified phosphorylation sites. (D) Immunoprecipitation assay using c-Myc antibody showing that FRQ failed to interact with Myc- CK-1a in the FRQ4A11 and FRQ4A12 mutants. (E) Western blot analysis showing that FRQ is hypophosphorylated in the FRQ4A11 and FRQ4A12 mutants. The asterisk indicates a nonspecific band recognized by our FRQ antibody. All cultures were grown in LL.