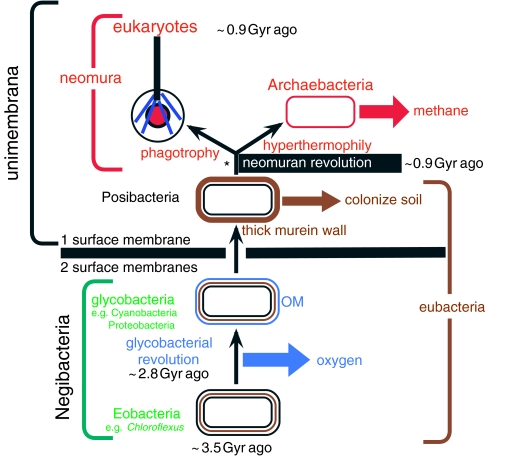
Figure 1.
The tree of life emphasizing major revolutions in cell structure. The most basic distinction in cell biology is between negibacteria, with a cell envelope of two distinct lipid bilayer membranes, and unimembrana, with but one surface membrane. Contrary to long-standing assumptions that cells first had only one membrane, phylogenetic analysis and palaeontology jointly show that the last common ancestor of all life was a negibacterium with two surface membranes (Cavalier-Smith 1987b, 2002a, 2006a), and that unimembrana evolved from them by losing the outer membrane (OM). Within negibacteria and unimembrana two revolutions in cell-surface chemistry created radically novel bacteria whose diversification transformed the biosphere and biogeochemical cycles. The glycobacterial revolution, soon after a photosystem duplication enabling oxygenic photosynthesis, complexified the OM by adding lipopolysaccharide and novel transport machinery. Nearly 2 Gyr later, the neomuran revolution made cell surfaces potentially more flexible by replacing the rigid eubacterial corset of cross-linked murein peptidoglycan by separate glycoproteins. The neomuran revolution was closely followed by the origin of the eukaryote cell; this entailed the origin of phagotrophy, the endomembrane system and endoskeleton and enslavement of a negibacterium (specifically an α-proteobacterium) to create mitochondria (not shown: see figures 5 and 6; Cavalier-Smith 1987b, 2002c, in press). At about the same time, life belatedly colonized hot, acid environments by evolving the ancestrally hyperthermophilic archaebacteria, sisters—not ancestors—of eukaryotes; distinctly later, one archaebacterial lineage evolved biological methanogenesis. The asterisk shows a widely assumed, incorrect position for the root of the tree that was based on a few paralogue trees for a subset of proteins that underwent an episode of such extensive quantum evolution during the neomuran revolution as to fabricate that misleading position of the root (between neomura and eubacteria) by a long-branch phylogenetic reconstruction artefact (Cavalier-Smith 2002a, 2006a). Metabolic enzyme paralogue trees typically place the root within Negibacteria, but in inconsistent places (Peretó et al. 2004).