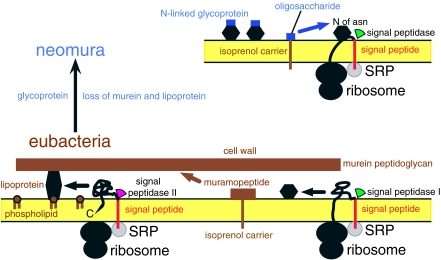
Figure 2.
Fundamental contrasts and continuities between eubacteria and neomura in surface chemistry and protein secretion. The older eubacteria (below) typically have walls of murein peptidoglycan and lipoprotein. Neomura have a surface coat (eukaryotes) or wall (archaebacteria) of N-linked glycoprotein (oligosaccharide covalently attached to protein via asparagine: Asn). Simple surface proteins in eubacteria (lower right hexagon) and protein parts of the conjugated glycoproteins and lipoproteins have hydrophobic N-terminal signal peptides recognized by homologous signal recognition particles (SRP: grey) that then link them to SRP-receptor proteins in the membrane and transfer the signal peptide into cross-membrane protein translocation channels (not shown). Ribosomes making surface proteins temporarily stick to the membrane at ribosome receptors, forcing the growing polypeptide chain through the channel. After synthesis, signal peptidases remove signal peptides of secreted proteins: signal peptidase I, used for simple eubacterial proteins, is homologous and ancestral to the neomuran signal peptidase. The very different signal peptidase II, specific for eubacterial lipoproteins, was lost during the neomuran revolution, as were lipoproteins and murein. Hydrophilic muramopeptide precursors of eubacteria and core oligosaccharides of neomura are moved across the membrane by long-chain amphipathic isoprenol carriers (dolichol in neomura, undecaprenol in eubacteria) to which homologous enzymes temporarily attach them via N-acetylglucosamine (GlcNAc; Cavalier-Smith 2006a) before being incorporated covalently into the wall. SRPs are more complex in neomura, with a novel protein, 19P, binding to an extra helix 6 in SRP RNA that is absent from the simpler, more primitive eubacterial SRP; this extra complexity stops proteins growing further until the ribosome/SRP complex correctly docks onto the membrane, an improvement to cotranslational protein secretion associated with novel cotranslational glycoprotein synthesis in the ancestral neomuran (Cavalier-Smith 2002a). For eubacteria, the condition in Posibacteria only is shown: the more complex negibacterial envelope has an OM outside the murein (figure 3). Neomura and eubacteria also differ substantially in DNA-supercoiling machinery: in both, DNA is similarly negatively supercoiled (i.e. underwound to help strands separate in transcription and replication), but eubacteria achieve this actively by the enzyme DNA gyrase, hydrolysing ATP, whereas neomura wrap DNA passively around core histones as nucleosome particles resembling beads on a string. Replacing DNA gyrase by histones had marked coevolutionary repercussions on DNA-handling enzymes, which were substantially modified during the neomuran revolution; archaebacteria alone invented reverse gyrase by fusing two eubacterial genes, a hyperthermophilic adaptation, showing they and hyperthermophily must be more recent (Cavalier-Smith 2002a).