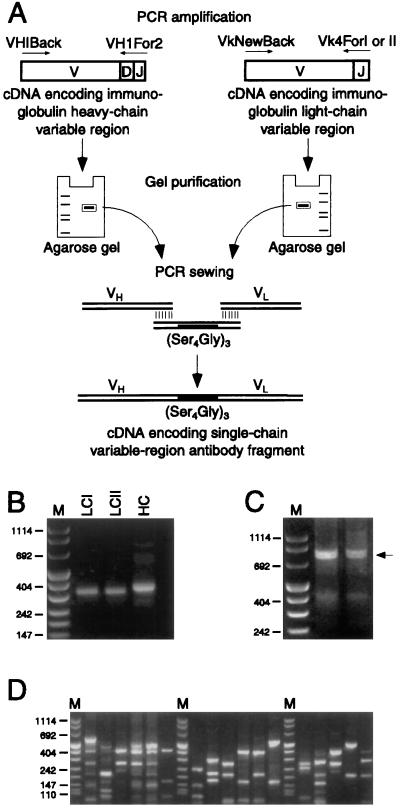
Figure 1.
Construction of the scFv insert and assessment of library diversity. (A) Lymphoid RNA, isolated from a mouse immunized with bullfrog sacculi and isolated hair cells, was reverse-transcribed and used as a template for PCR amplification of Ig heavy-chain (VH) and κ light-chain (VL) variable-domain repertoires. Heavy-chain primer VHIBack annealed to the 5′ end of the variable region (V) and VHIFor2 to the 3′ end of the joining region (J) of Ig heavy-chain cDNAs, resulting in products encoding the variable region, diversity region (D), and joining region of Ig heavy-chain proteins. Degenerate light-chain primers, VkNewBack and Vk4ForI or Vk4ForII, likewise annealed to kappa cDNAs, generating products encoding light-chain variable and joining regions. PCR products were gel-purified and randomly combined through an oligonucleotide encoding the (Gly4Ser)3 linker by splicing-by-overlap extension. Finally, restriction-endonuclease sites were appended to the assembled construct by the PCR and the purified product was ligated into an expression vector for bacteriophage display. This panel is based on an illustration in ref. 21. (B) Ig light-chain and heavy-chain variable domains were PCR-amplified from RNA present in the popliteal lymph nodes and spleen of a mouse immunized with bullfrog sacculi and isolated hair cells. Two pairs of primers were used to amplify the light-chain variable-domain repertoire; the amplified products generated by the VkNewBack and Vk4ForI primers are denoted LCI and the products generated by the VkNewBack and Vk4ForII primers are denoted LCII. The heavy-chain repertoire was amplified using the primers VHIBack and VHIFor2; the products are designated HC. (C) scFv fragments were constructed using PCR-sewing techniques to randomly join heavy-chain and light-chain variable domains. The sewn products of two reactions are indicated by an arrow. (D) Library diversity was ascertained by selecting bacterial colonies at random and examining their scFv insert sequences by PCR amplification and digestion with BstNI restriction endonuclease. Each lane contains the digestion products from the insert of a single bacterial colony; 17 representative clones are depicted here. This analysis indicates that the starting library was diverse, with no observed replication of digestion patterns. The lanes denoted M in B–D contain size markers of 1,114, 900, 692, 500, 404, 320, 242, 190, 147, 124, and 110 bp.