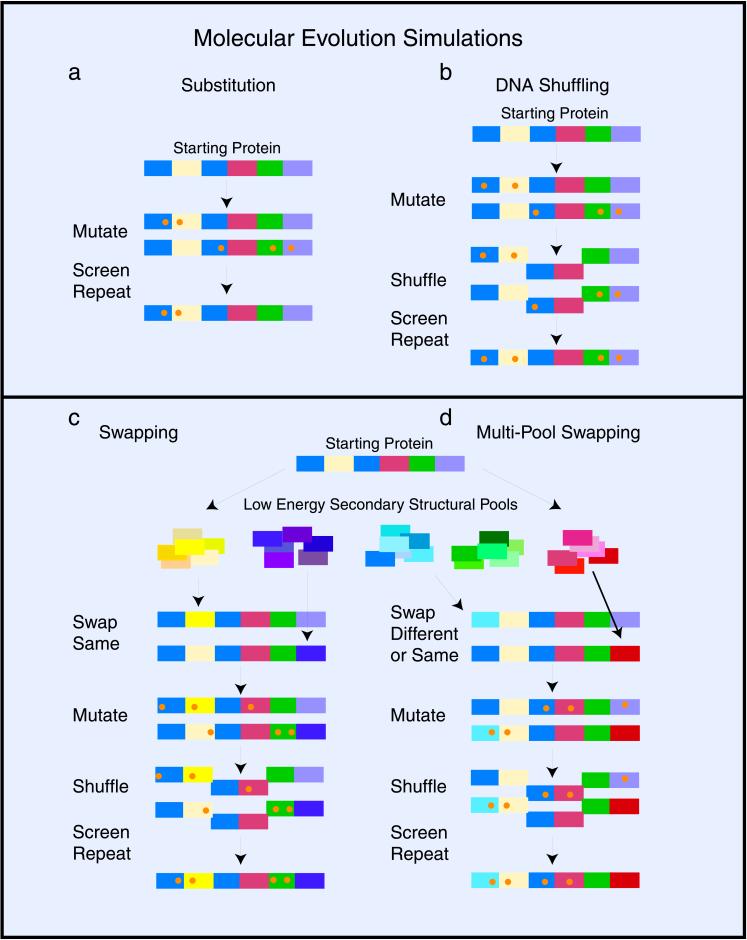
Figure 1.
Schematic diagram of the simulated molecular evolution protocols. (a) Simulation of molecular evolution by means of base substitution (substitutions are represented by orange dots). (b) Simulated DNA shuffling showing the optimal fragmentation length of two subdomains. (c) The hierarchical optimization of local space searching: the 250 different sequences in each of the five pools (e.g., helices, strands, turns, loops, and others) are schematically represented by different shades of the same color. (d) The multipool swapping model for searching vast regions of tertiary fold space is essentially the same as in the Fig. 1c, except that now sequences from all five different structural pools can be swapped into any subdomain. Multipool swapping allows for the formation of new tertiary structures by changing the type of secondary structure at any position along the protein.