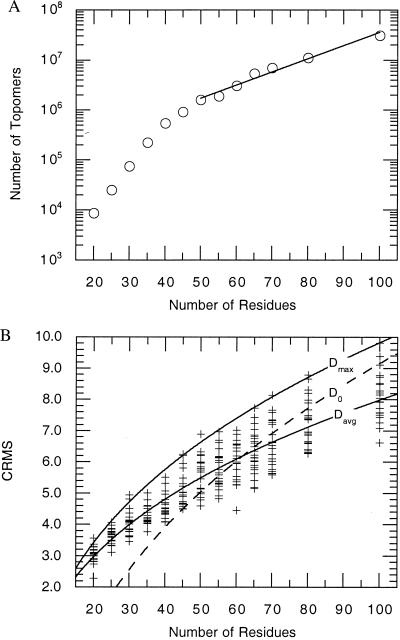
Figure 2.
(A) The number of disjoint topomers estimated for an N-residue polypeptide. Beyond N = 50, the number of topomers, SN, scales as SN = (83936) × (1.0624)N. For N = 100, the number of topomers is ≈(1.19)N. (B) The CRMS between each of the 277 native test conformations and their topomeric matches from the generic structure sets. The dashed line in the figure represents a previously developed average threshold for topological similarity developed by Maiorov and Crippen (11). They found that two N-residue structures are topologically similar when their CRMS is below the threshold, D0 = a + b (N)1/3, where a = −10.82 ± 0.37 and b = 4.31 ± 0.08. For N ≥ 50, the CRMS values we obtained from topomeric matches correlate well with the Maiorov-Crippen D0 threshold for topological similarity. Fitting a similar functional form to the average and maximum of our CRMS data for topomeric conformations yields Davg (a = −4.12 ± 0.24; b = 2.61 ± 0.06) and Dmax (a = −5.62 ± 0.40; b = 3.33 ± 0.11), respectively.