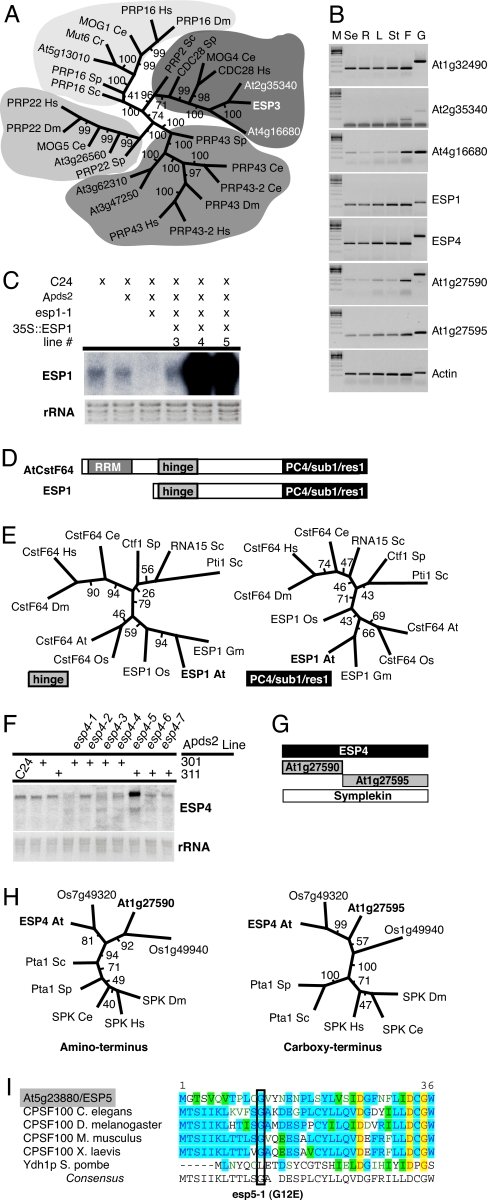
Fig. 2.
ESP genes encode RNA processing components. Bootstrap values for each internodal length corresponds to the frequency the two groups are separated in a population of 100 independent trees. Hs, Homo sapiens; Dm, Drosophila melanogaster; Ce, Caenorhabditis elegans; Sp, Schizosaccharomyces pombe; Sc, Saccharomyces cerevisiae; At, Arabidopsis thaliana; Cr, Chlamydomonas reinhardtii; Os, Oryza sativa; Gm, Glycine max. (A) Phylogenetic tree of ESP3 homologues by using the helicase domain and the neighbor-joining method. (B) Expression of ESP genes and closest Arabidopsis homologues (se, seedling; R, roots; L, leaves; St, stems; F, flowers; G, genomic DNA). (C) Northern blot analysis of ESP1 mRNA. (D) ESP1 is a CstF64 homologue. RRM, RNA recognition motif; Hinge, interacts with Symplekin and CstF77 in mammals; PC4/Sub1/Res1, interacts with polymerase processivity factors in mammals and yeast. (E) Phylogenetic trees of CstF64-related proteins based on the hinge and PC4/sub1/res1 domains (maximum parsimony). (F) ESP4 RNA levels. (G) There are three Arabidopsis Symplekin homologues. At1g27590 and At1g27595 have homology to the 5′ and 3′ halves of Symplekin and ESP4. RT-PCR detects expression from all 3 genes (see Fig. 2B). (H) Phylogenetic trees of ESP4 family members generated by a neighbor-joining method. (H Left) Amino terminus, amino acids 115–215 of ESP4. (H Right) Carboxyl terminus, amino acids 755–830. (I) Alignment of amino terminus of CPSF100 and esp5-1 substitution: red letters highlighted in yellow, absolute conservation; blue letters in aqua, conserved residues; black letters in green, similar residues; green letters (no highlighting), weakly similar residues.