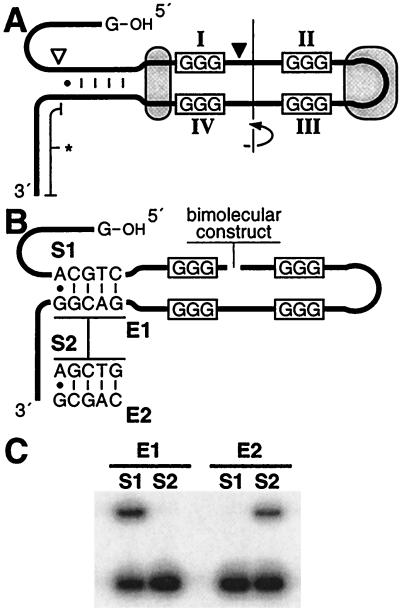
Figure 6.
Engineering an ATP-dependent deoxyribozyme. (A) A secondary-structure model for NTP-A2. Encircled regions identify highly conserved nucleotides. The 3′-terminal domain (∗) can be deleted with minor loss of activity. Filled and open arrowheads designate cleaved sites for the construction of bimolecular complexes. (B) Bimolecular constructs composed of separate NTP-A2.1 enzyme (E) and substrate (S) domains used to examine determinants of oligonucleotide substrate specificity. (C) PAGE analysis of the activity of E1 and E2 with the matched or mismatched substrates S1 and S2 that were internally 32P-labeled. Upper bands indicate deoxyribozyme activity. Substrate DNA (10 nM) was incubated with 100-fold excess of enzyme in the presence of 1 mM ATP for 5 hr before treatment with T4 DNA ligase.