Figure 7. Analysis of CPY sorting in wild-type and mutant Vps26p-expressing yeast strains.
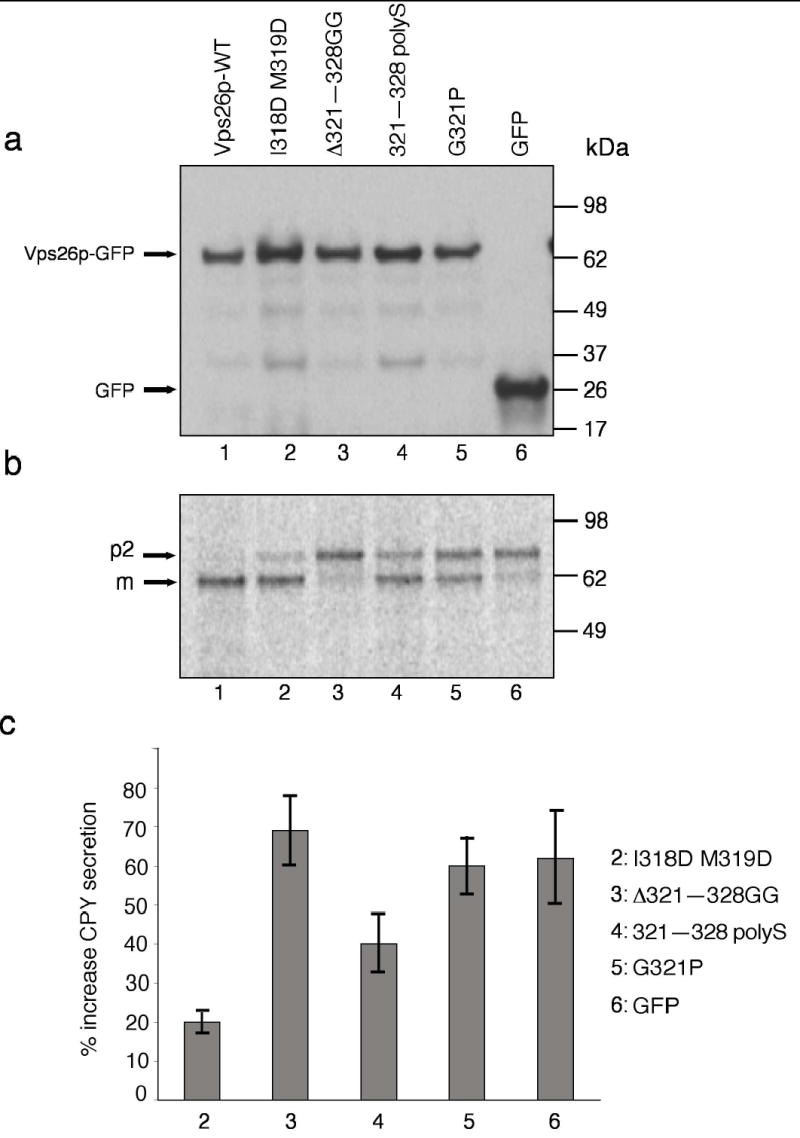
(a) S. cerevisiae vps26Δ strains transformed with expression plasmids encoding wild-type (lane 1) or I318D M319D (lane 2), Δ321–328 GG lane3, 321–328 polyS (lane 4) or G321P (lane 5) Vps26-GFP constructs, or GFP (lane 6), were lysed and analyzed by SDS-PAGE and immunoblotting using anti-GFP antibody. (b) The processing of CPY in the same strains described in (a) was analyzed by Metabolic-labeling, pulse-chase analysis and immunoprecipitation with antibody to CPY. Lanes correspond to the same constructs mentioned in (a). The positions of the Golgi precursor (p2) and mature (m) forms of CPY are indicated (c) Secretion of CPY into the medium by S. cerevisiae vps26Δ strains transformed with expression plasmids encoding the mutant Vps26-GFP constructs I318D M319D, Δ321–328 GG, 321–318 polyS and G321P, or GFP, was analyzed using a colony blotting assay. Values represent the percent increase of CPY secretion relative to the CPY secreted by a wild-type-Vps26-GFP-transformed vps26Δ strain. Results are expressed as mean ± SEM from four independent experiments.
