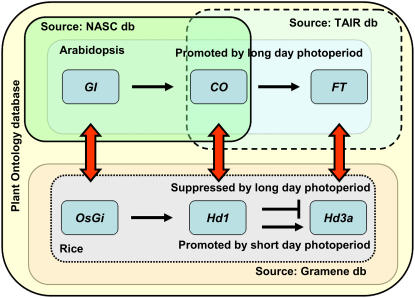
Figure 6.
Genes participating in the flowering time pathway. This image illustrates the flowering time pathway genes from Arabidopsis, GI, CO and FT, and rice, OsGI, Se1 (Hd1), and Hd3a. In the PO database, the annotation for these genes is provided by three databases, the National Arabidopsis Stock Centre, TAIR (for Arabidopsis), and Gramene (for rice). The curators have used terms (Table III) from the whole GSO and PSO to suggest when and where in a plant these genes were expressed or their phenotype was observed. Based on the experiment types (evidence codes) and citation evidences, the databases recorded information about the mutant/gene/gene product to the GSO and the PSO terms. Compared to the short-day length promotion of flowering in rice, flowering is promoted by long-day exposure in Arabidopsis. When rice is exposed to long days, it leads to a down regulation of the Hd3a gene by Se1 (Hd1), leading to a delayed transition of the vegetative shoot apical meristem to the reproductive inflorescence meristem. In other words, the growth stage inflorescence visible (sensu Poaceae), which is synonymous with heading stage, is delayed. The double-headed arrows suggest that the Arabidopsis and rice genes are orthologous. The colored boxes around the genes represent the databases that provided the gene annotations. In the PO database, the putative orthology of these genes cannot currently be determined or displayed, but it can be inferred by visiting either the Gramene or the TAIR database.