FIG. 4.
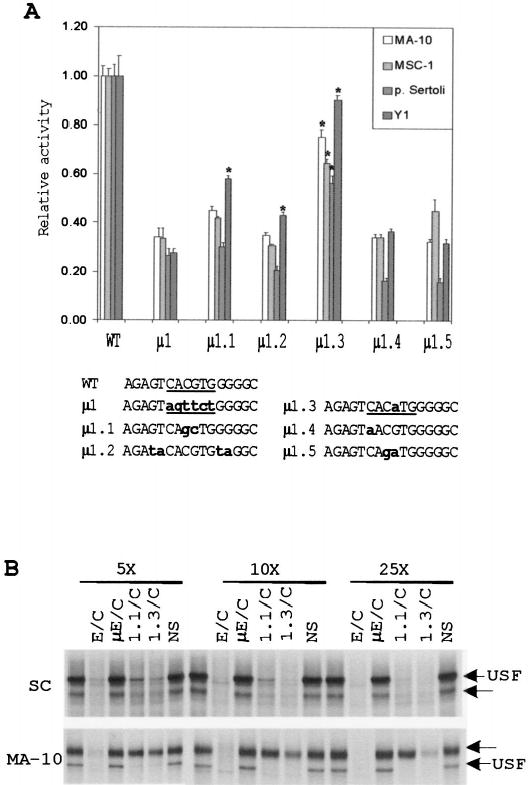
Point mutations in the E box reveal bases within the core and flanking sequences that are needed for its function. A) Five single- or double-base substitutions of the E box were generated in the context of the SF1(− 734/+60)Luc reporter plasmid. The constructs were transiently transfected into MA-10, MSC-1, primary Sertoli cells, and Y1 cells, and their activities were determined by the Dual-Luciferase Reporter Assay (see Materials and Methods). All transfections were performed in the presence of pRL-TK (Renilla luciferase expressed from the viral thymidine kinase promoter), which was used as a control for transfection efficiency. The relative activity represents the average firefly/Renilla luciferase values of each mutant relative to the firefly/Renilla luciferase activity of the wild-type SF1(− 734/+60)Luc construct. Data represent a minimum of three independent transfections for each cell type. Error bars represent the SEM. Sequences of the mutants from positions − 87 to− 72 are shown at the bottom. The E boxes are underlined, and bold lettering indicates the mutated nucleotides. Asterisks indicate values that are statistically different from those of mutant μ 1 for each cell type according to the Student t-test (assuming unequal variance with P < 0.01). B) EMSA was performed using a radiolabeled E box probe together with nuclear proteins extracted from primary Sertoli cells and MA-10 cells as described in Figure 2. To determine the relative affinities of the retarded E box complexes, a 5-, 10-, and 25-fold molar excess of various competitors (indicated above the lanes) was added to the binding reaction. The structures of the E box probe and competitor oligodeoxynucleotides used are given in Table 2. Arrows indicate positions of two specific complexes.
