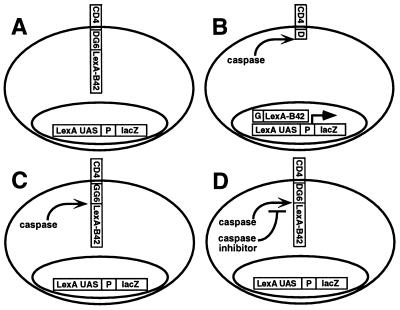
Figure 1.
A genetic system for monitoring caspase activity in yeast using a transcriptional reporter. Yeast were created that express a chimeric type-1 transmembrane protein (CLBDG6) in which the N-terminal signal sequence and transmembrane domain (CD4) is followed by a linker consisting of six tetrapeptide caspase target sites (indicated in bold) that bracket the specificity of known caspases and granzyme B (23)—DEVDG-WEHDG-IEHDG-IETDG-DEHDG-DQMDG—each of which is followed by a glycine residue, which acts as a stabilizing residue in the N-end rule degradation pathway in yeast (reviewed in ref. 24). C-terminal to the caspase target site linker is a transcription factor domain, LexA-B42. The LexA-dependent transcriptional reporter consists of LexA binding sites (LexA UAS) and a promoter (P) upstream of the bacterial lacZ gene (lacZ) (A). The cells in A act as caspase activity reporters because expression of an active caspase results in CLBDG6 cleavage at the caspase target sites, releasing LexA-B42, which enters the nucleus and activates lacZ transcription (B). A version of CLBDG6 in which the P1 aspartates are changed to glycines (CLBGG6) cannot be cleaved by caspases. Cells expressing CLBGG6 act as false positive reporters for molecules that activate lacZ expression independent of cleavage at caspase target site (C). If cells in B express a caspase inhibitor as well as an active caspase, caspase activity, and thus caspase-dependent release of LexA-B42, is inhibited, and β-gal levels are decreased compared with cells that express the caspase alone (D).