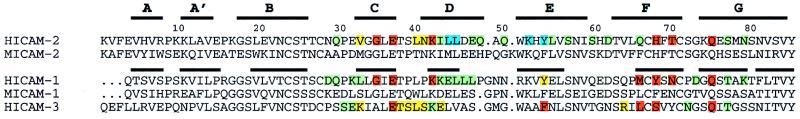
Figure 2.
LFA-1 binding residues in the ICAM subfamily of adhesion receptors. N-terminal domains of ICAM-1 and -2 were structurally aligned with 3dmalign of modeller with a gap penalty of 1.75 Å (12), and ICAM-3 was aligned based on sequence homology. Thick lines represent β-strands in the structures of ICAM-2 and ICAM-1. Residues found important for LFA-1 recognition by single residue mutagenesis experiments as described here for ICAM-2 or elsewhere for ICAM-1 or ICAM-3 (7, 20–23, 25) are color-coded according to percentage of wild-type binding: red, severe, <35%; orange, moderate, 35–70%; green, little or no effect, 70–100%; cyan, <35% binding with a possible effect of the mutation on structural integrity. Mutation was to alanine, except for mutations Y52/F (7) and Y66/S (20) in ICAM-1, G35/T in ICAM-2, and mutations L66/K and S68/K in ICAM-3 (21).