Figure 1.
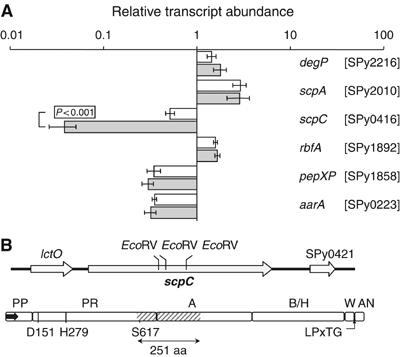
Identification of ScpC as a putative IL-8 protease. (A) The effect of SilCR on the transcription of GAS serine peptidase. The abundance of the indicated serine peptidase transcripts relative to that of gyrA was determined by real-time RT–PCR in RNA derived from WT grown to OD of 0.4 at 600 nm in the absence (clear bars) or presence (shaded bars) of SilCR (10 μg/ml). The values are mean obtained from analysis in duplicate of three independent RNA samples. Error bars represent standard deviation (s.d.). SilCR downregulates the transcription of scpC. P<0.001 (Student's test). (B) Genomic arrangement and domain organization of ScpC. Upper panel: The arrows depict the identified ORFs and their direction of transcription. The 5′ and 3′ EcoRV sites at positions 1801 and 2554 bp from the beginning of scpC were used to replace the internal scpA coding sequence with Ωkm2. spy0421 encodes a product of 236 aa with no homology to characterized proteins. Lower panel: The map of the motifs identified in the predicted sequence of ScpC includes the following: the pre-pro (PP) domain (residues 1–123) containing the signal sequence (residues 1–34) depicted as an arrow, protease domain (PR) (residues 124–688) containing Asp, His, and Ser forming the catalytic triad; the A domain (residues 689–1128) and the B/H domain (residues 1129–1560), the cell wall domain (W) (residues 1561–1613), the cell wall anchor domain (AN) (residues 1613–1647), and the LPxTG motif starting at residue 1613. The DNA region removed by the EcoRV digestion contains a segment of 251 aa (hatched) including Ser617 of the catalytic triad.
