Figure 4.
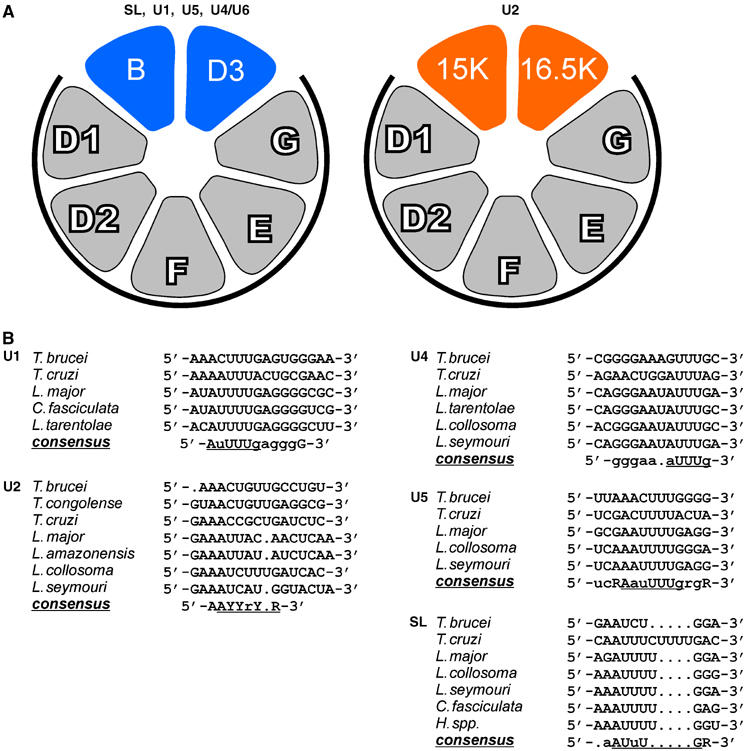
Model of Sm core variation in the U2 snRNP (A) and sequence comparison of Sm sites in trypanosomid snRNAs (B). (A) The arrangement of Sm proteins in the standard core (left model) is according to Palfi et al (2000). This composition of Sm polypeptides has been experimentally verified for the SL RNP, and the U1, U5, and U4/U6 snRNPs from T. brucei (this study; see Figure 2). The U2 snRNP, however, contains a variant Sm core, in which the canonical SmB and SmD3 proteins are replaced by Sm15K and Sm16.5K (right model). The relative orientation of the latter two Sm proteins is based on their homology to SmB and SmD3, respectively, and on protein–protein interaction data (see Figure 3). The common subcore of SmD1/D2/F/E/G is marked by heavy lines. (B) Sequence alignment of the Sm sites of U1, U2, U4, U5 snRNAs as well as of the SL RNA from the following trypanosomatid species: T. brucei; T. cruzi, T. congolense, L. major, L. amazonensis, Leptomonas collosoma, Leptomonas seymouri, Crithidia fasciculata, Herpetomonas species. For U1, U4, U5, and SL RNAs, sequences from only a few representative species are shown, for U2 all available sequences. Nucleotides in large letters indicate absolute conservation for the sequences shown, small letters indicate at most one deviation. The consensus sequences of the Sm sites are underlined. R, purine, Y, pyrimidine nucleotide.
