Figure 1.
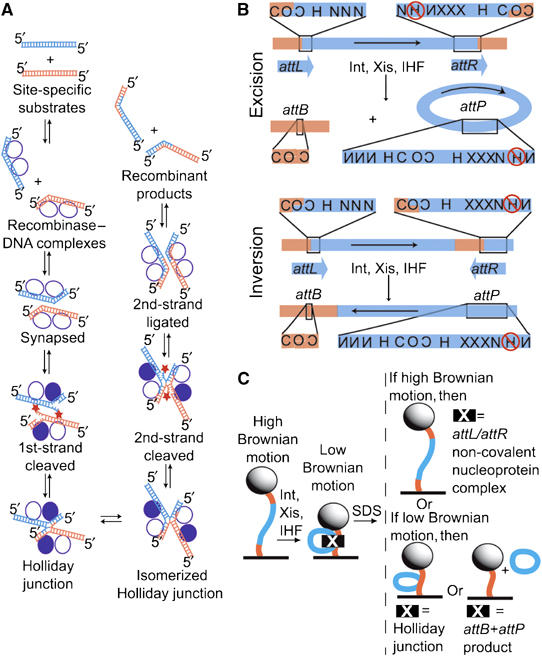
Intramolecular recombination substrates, generated products, and experimental design. (A) General scheme of the site-specific recombination reaction, in which parental DNAs (orange and blue) are recombined to form two hybrid DNA products (see text for details). The recombinase subunits (circles) that catalyze these reactions form a tetramer in which only one pair (filled) is DNA cleavage active. The reactivity of the Int pairs is reversed for the second pair of strand exchanges. All strand exchanges proceed through two successive transesterification reactions: a strand cleavage reaction in which a tyrosine side chain attacks the DNA backbone and a ligation in which the resulting phosphotyrosine linkage is attacked by a free DNA 5′ hydroxyl (star). (B) Maps of the recombination substrates and the products generated from their recombination. Top (excision): when the phage λ attachment sites are in direct repeat (attL+attR substrate), recombination generates separated linear attB and circular attP products. Bottom (inversion): the indirect repeat substrate (attLinvattR) recombines to form a single linear product in which the DNA between attB and attP attachment sites has been inverted. The presence of Xis prevents further recombination of both the excision and inversion products (Abremski and Gottesman, 1982). att sites contain 7 bp homologous DNA sequences (overlap region, O) between cleavage sites and multiple sites for binding of protein domains: N, Int N-terminal domain; C, Int catalytic domain; H, IHF; and X, Xis. Reversed lettering indicates inverted site orientation. IHF binding to the circled site inhibits excision; to avoid this, this site was inactivated by mutations (Thompson et al, 1986). The binding site for Fis protein, which overlaps with one of the Xis sites, is not shown because this host-encoded DNA-bending protein is not required for excision in the presence of sufficient Xis (Thompson et al, 1987a, 1987b) and was not included in these experiments. Both substrates are 1943 bp long and have att site overlap regions positioned 301 and 290 bp from the closest end. (C) Experimental design with attL+attR DNA. Each 200 nm polystyrene bead (sphere) is tethered to the coverslip surface of a flow chamber (black line) by a single substrate DNA molecule (orange and blue curve; Finzi and Gelles, 1995). Formation of non-covalent protein–DNA complexes that bend or loop DNA (e.g., att-site complexes or synaptic complexes) reduce the effective length of the tether and can be detected as a decrease in bead Brownian motion. Formation of covalent links between two DNA segments (in a Holliday junction) or excision of a DNA segment will have a similar effect. Effective tether length changes resulting from non-covalent protein–DNA interactions and those from covalent DNA backbone modifications can be distinguished by challenge with the protein denaturant sodium dodecyl sulfate (SDS).
