Figure 2.
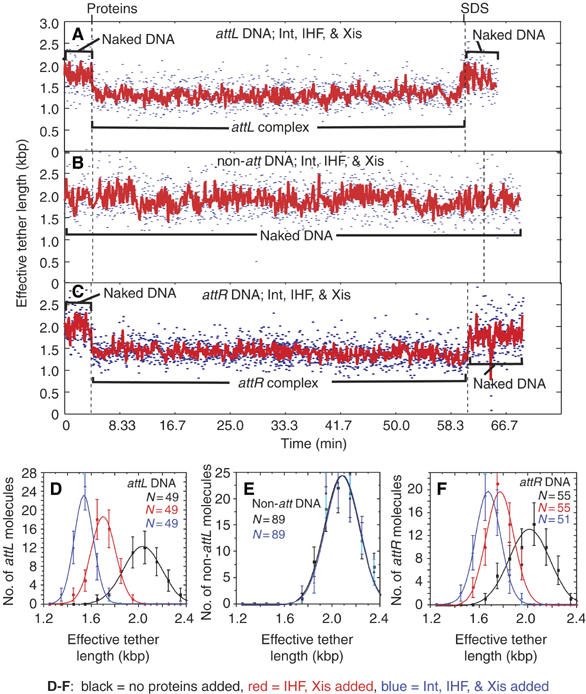
Specific complex formation on DNA molecules with only one att site. (A–C) Changes in the effective lengths of single DNA molecules in response to the addition of recombination proteins Int, Xis, and IHF. The three proteins or the SDS challenge were added at the times indicated by dashed lines. Both unfiltered (blue; 2.13 s time resolution) and filtered (red) data are shown. Noise in tethered-particle motion (TPM) measurements increases with the increasing tether length (Yin et al, 1994). Wild-type proteins were added to single DNA molecules containing a single attL (A), no att sites (B), or a single attR (C). (D–F) The distributions (±s.e.) of effective tether lengths (and Gaussian fits) compiled from all experiments for attL-containing molecules (D), non-att DNA molecules (E), or attR-containing DNA molecules (F). The measurement for each type of molecule was calculated as the mean of the 10.5 s of tether length data: commencing at t=0 min (before protein addition; black squares), commencing at t=7.5 min (after IHF and Xis addition; red circles), or commencing at t=7.5 min (after wild-type Int, Xis, and IHF addition; blue triangles).
