Figure 2. Targeted mutagenesis of the F. nucleatum ATCC 23726 chromosomal rnr gene.
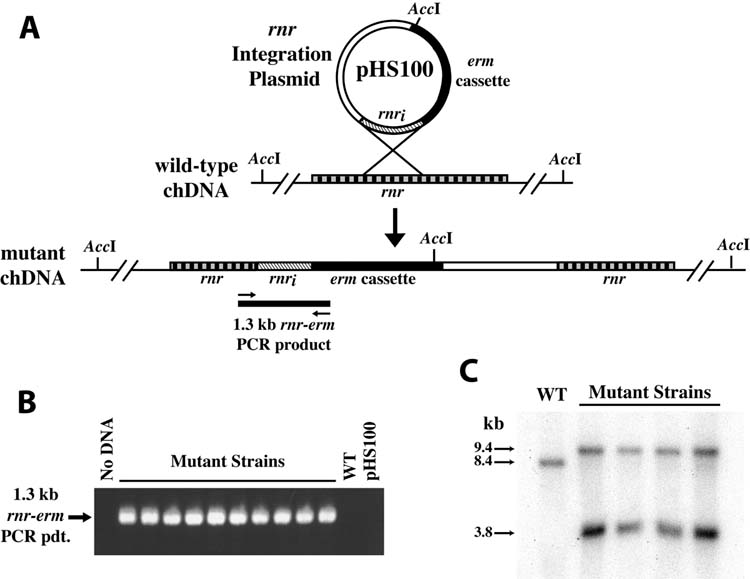
The rnr integration plasmid pHS100 was constructed with a fragment of the rnr gene to provide a region of DNA enabling homologous recombination of the suicide plasmid into the F. nucleatum chromosomal rnr gene (Panel A). Analysis of the mutants (Panels B and C) was consistent with this prediction. A: Schematic diagram of the integration plasmid pHS100, wild-type and mutant chromosomal structure. The position of primers and the predicted PCR amplicon specific to the chromosomal insertion of pHS100 are indicated. The position of the primers (SA1 and SA2) and the predicted 1.3 kb amplicon are indicated. Note also integration of the plasmid introduces a unique AccI site between the 5′ and 3′ ends of the rnr gene. B: PCR analysis revealed the presence of the 1.3 kb amplicon specific to the chromosomal insertion with the mutant strain chromosomal DNA. As predicted, no PCR product was evident in the absence of DNA template (No DNA), or when using either the parental chromosomal DNA (WT) or pHS100 as template (IP). C: Southern blot of AccI-digested chromosomal DNA from the parental strain ATCC 23726 (WT) and representative mutant strains was probed with a radiolabeled rnr DNA probe. In the parental wild type chromosomal DNA a single AccI band is detected with the rnr probe, as predicted due to the lack of an AccI site in the wild type rnr gene. In contrast, two hybridizing DNA bands are evident in the chromosomal DNA of the mutant strains, consistent with the introduction of a unique AccI site present in the rnr integration plasmid. The approximate molecular mass of the bands is indicated on the left.
