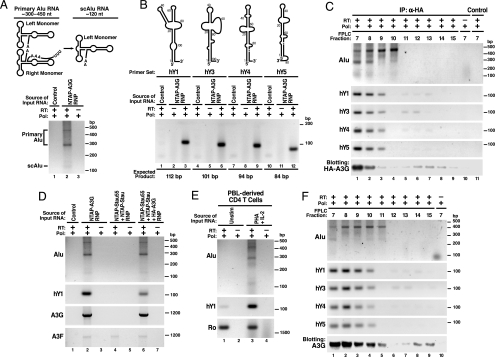
Fig. 4.
Detection of nonautonomous mobile genetic elements in the HMM A3G complexes. (A Upper) Schematic of primary Alu and processed scAlu RNAs. (A Lower) RT-PCR detection of primary Alu and scAlu RNAs in HMM A3G complexes. Control purifications: unlinked NTAP+HA–A3G. (B Upper) Schematic of hY RNAs. (B Lower) RT-PCR detection of hY RNAs in HMM A3G complexes. (C) Co-IP of Alu and hY RNAs with HA–A3G from HMM fractions of 293T lysates resolved by FPLC. Protein G-agarose in the absence of anti-HA served as a control. (D) A3G-dependent recruitment of Alu and hY RNAs into the Staufen-containing RNA granules. A3G and endogenous A3F mRNAs are also present in purified NTAP–A3G and NTAP–Stau (Stau55) complexes. Of note, 293T cells expressed extremely low levels of A3F. Alu RNA was not present in the RNA granules unless A3G was coexpressed. (E) PHA–IL-2-induced expression of Alu and hY RNAs in primary CD4 T cells. Cells were either untreated or treated with PHA (5 μg/ml) for 36 h followed by IL-2 (20 units/ml; Roche) for 36 h before analysis. (F) Alu and hY RNAs cofractionate with HMM A3G complexes in PHA–IL-2 activated primary CD4 T cells. Reactions performed with Pfx polymerase (Pol) but not reverse transcriptase (−RT) served as negative controls in each panel. RNA structures in A and B were adapted in part from refs. 45 and 46.