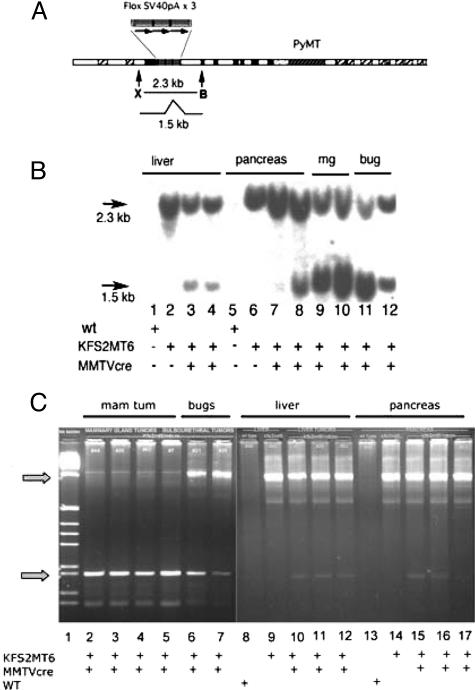
Figure 5.
Cre-dependent transgene recombination. A: Map of KFS2MT transgene showing a 2.3-kb XhoI (X) to BglII (B) fragment that is shortened to 1.5 kb by Cre-mediated deletion of triplet-repeated SV40Pa elements. B: Southern blot analysis of KFS2MT6 alone and in combination with MMTV-Cre7 transgene as indicated at bottom of panel. DNAs isolated from liver, pancreas, mammary tumors (mg), and bulbourethral gland (bug) were digested with XhoI and BglII and detected by hybridization with a 250-bp probe (AF179904: 2576–2825) synthesized by PCR. The recombined (1.5 kb) form of the gene was detected in lanes 3, 4, and 7 to 12. Note the strength of the signal of the 1.5-kb form of the mammary tumors of lanes 9 and 10 are similar to the monogenic control of lane 6. C: Genomic PCR confirmation of Cre-dependent recombination. Genomic DNA isolated from the indicated tumors or organs was subjected to PCR with primers flanking the floxed stop signals. This resulted in a larger fragment (arrow) of 1 kb (top arrow) for a nonrecombined transgene or a 0.2-kb fragment (bottom arrow) for a Cre-mediated recombination event. Note the abundance of the 0.2-kb PCR product in mammary tumors (mam tum) and bulbourethral gland tumors (bugs) and weak lower band in KFS2MT6; MMTV-Cre7 bigenic liver and pancreas.