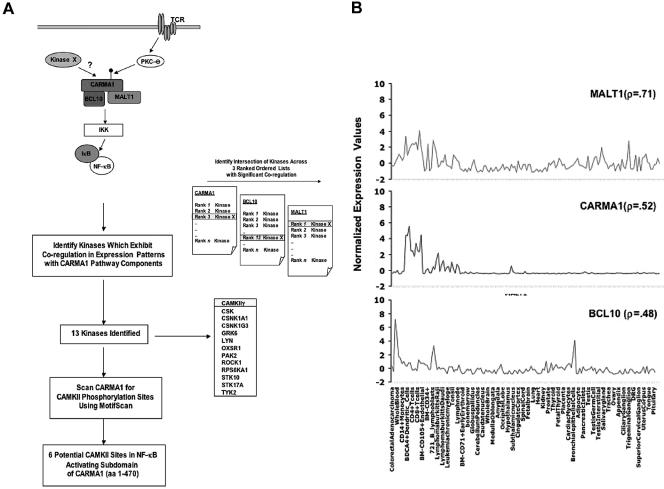
FIG. 1.
Schema for identification of CaMKII as a potential modulator of CARMA1-mediated NF-κB activation by use of kinase and pathway component expression signatures. (A) (i) Literature analysis of pathway components in TCR-mediated NF-κB activation identified CARMA1, Bcl10, and MALT1 as a functional module whose components exhibit tissue-restricted expression profiles. We hypothesized that an additional CARMA1 regulatory kinase (kinase X) would exhibit a coregulated expression signature with CARMA1, Bcl10, and MALT1. (ii) We derived a list of potential candidate kinases based on expression profiles. Three rank-ordered lists of kinase expression profiles with respect to CARMA1, Bcl10, and MALT1 gene sets were generated. For CARMA1, MALT1, and Bcl10, each of the probes representing the 460 kinases in the data set are ranked from most similar to each to least similar in expression profile across the GNF compendium based on pairwise Pearson's correlation coefficient. Subsequently, 13 kinases were identified among 460, including CaMKIIγ, which exhibited significant correlation with the target gene set of the CARMA1 signaling pathway. (iii) Motif Scan was employed to examine if CARMA1 had potential consensus CaMKII phosphorylation sites. (B) Expression level correlation between CaMKIIγ and the CARMA1-MALT1-Bcl10 gene set. A Z-score normalization (y axis) of microarray intensity values was performed and plotted for CaMKIIγ and the CARMA1-MALT1-Bcl10 target gene set in each of the tissue/cell types (x axis) in the GNF compendium data set (all 79 tissues/cells in tissue compendium are plotted, and a selected subset is labeled). Also indicated are the Pearson's pairwise correlation values (r) between CaMKIIγ and the individual gene set members as a quantification of expression similarity. DRG, dorsal root ganglion.