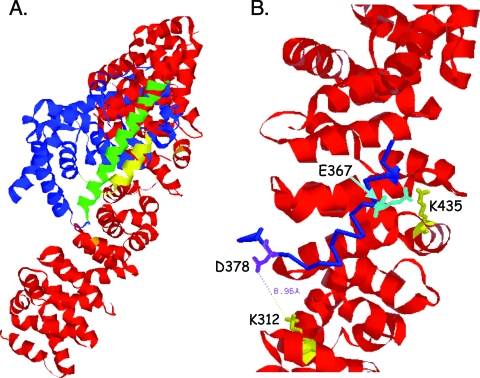
FIG. 7.
Predicted three-dimensional model of PPARγ interacting with β-catenin. (A) The ribbon model shows the entire structure of PPARγ (blue) and β-catenin (red). The model was generated using the protein-protein docking program (ClusPro) available at http://nrc.bu.edu/cluster. PDB codes 2prg (for PPARγ) and 1LUJ (for β-catenin) were downloaded from the PDB website (http://www.pdb.org). The yellow region (helices 7 and 8, aa 367 to 382) of PPARγ is shown in close proximity to the charged buttons, K435 to K312, of β-catenin and appears not to interfere with helix 10 (green), which is required for binding to RXRα. (B) A region of the model from panel A was enlarged to show the interaction of amino acids 367 to 382 of PPARγ (blue, backbone only shown) with the charged buttons K312 and K435 (yellow) of β-catenin (red). The side chains of two positively charged amino acids (K312 and K435) of β-catenin and two negatively charged amino acids (D378 in purple and E367 in cyan) of PPARγ are shown in stick drawing. The distance between the K312 (β-catenin) and D378 (of PPARγ) side chains is 8.96 Å, which is within the distance to form a “salt bridge.”