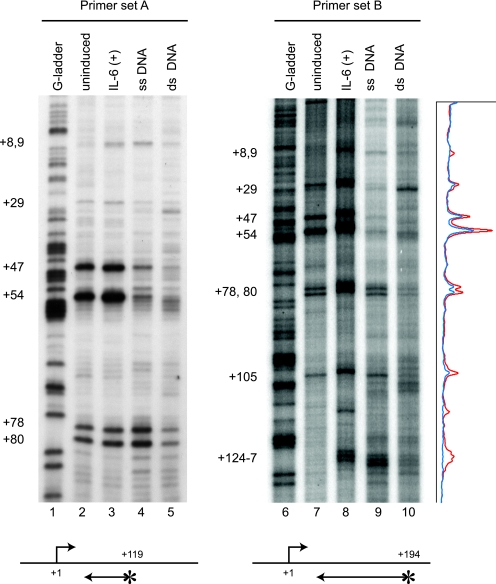
FIG. 3.
RNAPII strongly accumulates at around position +50 of the junB gene. Control cells or cells stimulated by IL-6 for 15 min were treated with 7.5 mM KMnO4. Genomic DNA was purified, and unpaired thymine residues in transcription bubbles were mapped by in vivo footprinting with two primer sets. KMnO4 sensitivity in vivo was compared to the sensitivity of purified single-stranded (ss) or double-stranded (ds) genomic DNA. Lanes 1 and 6 contain genomic DNA partially cleaved at guanine residues. Footprinting with primer set A (lanes 1 to 5) results in higher-resolution mapping of paused sites in a smaller region than that with primer set B (lanes 6 to 10). Signal intensity of the footprinting data for primer set B was obtained with a Storm 860 image analyzer and is presented on the right. The blue and red lines represent the results of uninduced and induced states, respectively. The positions of end-labeled primers are indicated by arrows with asterisks. The numbers on the left indicate the relative positions from the transcription initiation site.