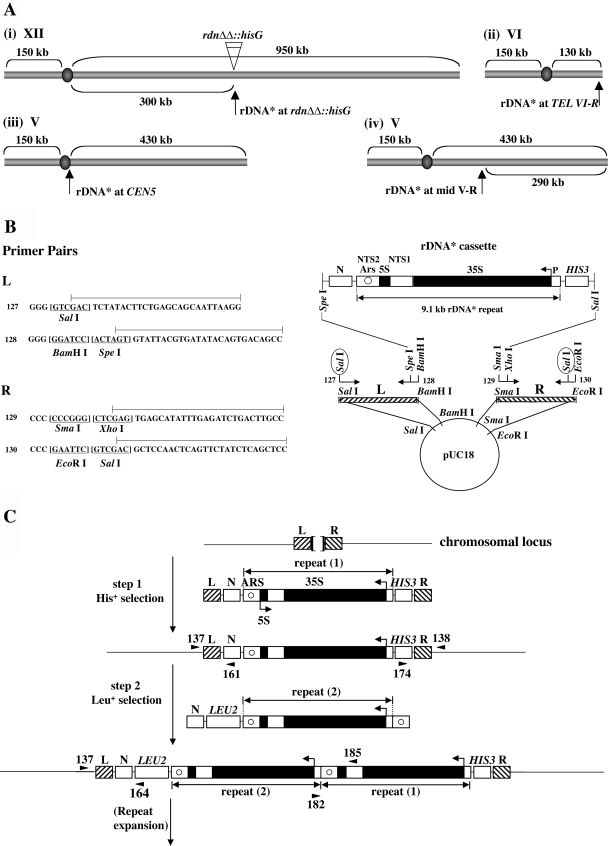
FIG. 1.
Integration of and expansion of a new rDNA repeat at both normal and ectopic chromosomal sites. (A) Sites of integration carried out in the present study. A new rDNA repeat carries a hyg1 mutation and a 26-bp tag (see the text and Fig. 2A) and is designated “rDNA*.” (B) Construction of plasmid pNOY3292 used for integration of rDNA* at the rdnΔΔ::hisG locus [see Fig. 1A(i)]. The rDNA* cassette consists of an entire 9.1-kb rRNA gene copy flanked by the HIS3 gene and a non-yeast sequence N from A. thaliana. The 35S and 5S rRNA coding regions are shown as filled regions of the rDNA* repeat. Nontranscribed sequences 1 and 2 are labeled as NTS1 and NTS2, respectively. ARS is shown as a circle within NTS2. The lines above the primer sequences (primers 127, 128, 129, and 130) show the regions that are identical to the sequences of chromosomal DNA flanking the RDN1 locus (called “L” and “R”). Sequences of restriction enzyme sites are underlined. Digestion of plasmid pNOY3292 with SalI (circled) will produce an ∼13-kb fragment containing L, N, rDNA*, HIS3, and R. This fragment was used for step 1 shown in C. (C) General strategy used for the integration of rDNA* at a given chromosomal site and its subsequent expansion. Two separate steps and DNA fragments used for integration of rDNA* are shown. The sequences flanking the site of integration are shown as L and R. For integration of rDNA* at the original RDN1 site in rdnΔΔ::hisG strains, the Escherichia coli hisG sequence is between L and R flanking sequences (as shown by a bracket at the top of the figure). In the other three cases (see panel A), the sequences between L and R are 0 to 8 bp as described elsewhere (see the supplemental material). The DNA fragment used for the second step is an ∼13-kb fragment obtained after digestion of pNOY3293 with SpeI and SalI. The positions of primers used to confirm correct integration are shown as arrowheads, taking examples for integration at the rdnΔΔ::hisG locus. The primer pairs 137/161 and 174/138 produce 1,036- and 1,263-bp fragments, respectively, for correct integration at the first step. PCRs using primer pairs 137/164 and 182/185 produce 2,007- and 1,800-bp fragments, respectively, for correct integration at the second step. For integration at other ectopic loci, primers at equivalent positions were used for PCRs as described elsewhere (see the supplemental material).