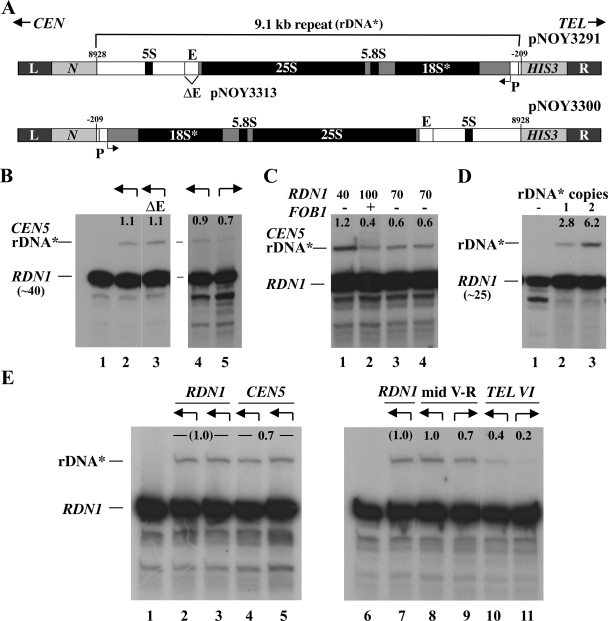
FIG. 3.
Transcription of a single rRNA gene at an ectopic site in the presence of reduced copy number of rRNA genes at RDN1. (A) Structures of the DNA fragment carried by pNOY3291 used for integration of a single copy of rDNA* near CEN5 and that carried by pNOY3300 used for integration of a single-copy rDNA* in the opposite orientation near CEN5. The L and R sequences are those flanking the sites of integration, as explained in Fig. 1B and C. The structure of the first DNA fragment is the same as that shown in Fig. 1B, with more details identified. The Pol I transcription start site is defined as +1, and the position of the E element (the 319-bp EcoRI-HpaI region) is indicated. pNOY3313 is the same as the pNOY3291 used for integration near CEN5 of rDNA* but has a deletion of the E element (ΔE; see lane 3 in panel B). (B) RNA samples (5 μg) from NOY2063, NOY2064, and NOY2147 (lanes 1 to 3, respectively) were analyzed by primer extension. In a separate experiment, RNA samples (10 μg) from NOY2064 (lane 4) and NOY2146 (lane 5) were similarly analyzed. The amounts of transcripts from ectopic rDNA* and those from the 40 copy repeats at RDN1 were measured by using a PhosphorImager, and the ratios of the ectopic ones to those from the RDN1 are given as percentage values above the rDNA* bands. The arrows above each lane indicate the direction of transcription of the rRNA* gene integrated at the CEN5 ectopic site as shown in panel A. (C) RNA samples (10 μg) from NOY2064 (lane 1), NOY2064 after introduction of FOB1 (lane 2), and after introduction followed by the removal of FOB1 (lanes 3 and 4 for two independent isolates) were analyzed by primer extension. The rDNA copy numbers at RDN1 of the same cultures used for primer extension were measured by Southern blot analysis and are indicated above the lanes. The expression of rDNA* relative to that of rDNA repeats at RDN1 was calculated and the values are indicated above the rDNA* bands as in panel B. (D) An autoradiogram showing transcripts of rDNA* compared to RDN1 (∼25-copy strain). RNA samples (5 μg) from NOY2068 (lane 1), NOY2159 (lane 2), and NOY2158 (lane 3) were analyzed by primer extension. The expression of rDNA* relative to that of rRNA gene repeats at RDN1 was calculated, and the values are indicated above the rDNA* bands as in panel B. The results shown are from one of several independent transformants. The values from this and other experiments averaged 2.8 ± 0.2 for NOY2159 and 6.2 ± 0.4 for NOY2158. (E) Transcription of a single rRNA* gene at an ectopic site compared to the expression of a single rDNA* copy integrated into native copies at RDN1. Arrows above each lane indicate the directions of transcription of the rDNA* gene shown in the way similar to that shown in panel B. Autoradiograms show the transcription of rDNA* analyzed by primer extension. RNA samples (5 μg) from NOY2063 (lane 1); NOY2148 (two independent rDNA* integrants [lanes 2 and 3]); NOY2064 (two independent rDNA* integrants [lanes 4 and 5]); and NOY2063, NOY2148, NOY2149, NOY2150, NOY2151, and NOY2152 (lanes 6 to 11, respectively) were analyzed. The ratios of expression of rDNA* to that of the native rRNA gene repeats (∼40 copies) at RDN1 were measured. The average of values for lanes 4 and 5 were normalized to the average of those for lanes 2 and 3. The average of the normalized value from this and other experiments was 0.68 ± 0.09 and is shown as 0.7. For lanes 8 to 11, the ratios of rDNA* to rRNA gene repeats at RDN1 transcripts were measured and normalized to the value for NOY2148 (lane 7). Two independent integrants were used for each ectopic site and both orientations. The normalized values from this and other experiments were averaged, yielding values for samples corresponding to lanes 8, 9, 10, and 11 to be 1.03 ± 0.17, 0.68 ± 0.21, 0.40 ± 0.17, and 0.21 ± 0.17, respectively. The rounded numbers are given above the rDNA* bands.