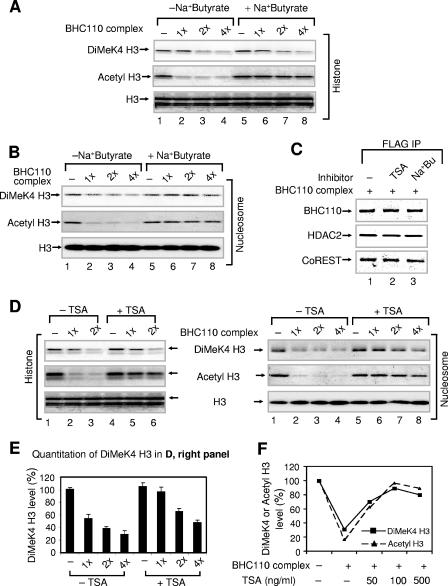
FIG. 4.
Inhibition of deacetylation reduces nucleosomal demethylation. (A) Effect of sodium (Na+) butyrate (100 mM) on histone demethylation. Bulk histones were used as substrates. (B) Effect of sodium butyrate (100 mM) on nucleosomal demethylation. Nucleosomes were used as substrates. “1×” corresponds to approximately 50 ng of HDAC1/2 in the complex. (C) Immunoprecipitation of the BHC110 complexes in the absence or presence of trichostatin A or sodium (Na+) butyrate. The BHC110 complexes were incubated without or with TSA (500 ng/ml) and sodium butyrate (Na+Bu, 100 mM), immunoprecipitated, and analyzed by Western blotting. (D) Effect of TSA (500 ng/ml) on histone (left) and nucleosomal demethylation (right). Bulk histones and nucleosomes, respectively, were used as substrates. (E) Quantitation of dimethyl K4 H3 levels shown in panel D, right panel. The dimethyl K4 H3 levels in the absence of the complex (−) were set as 100%. Data are means plus standard deviations from three different experiments. (F) Effect of different concentrations of trichostatin A on nucleosomal demethylation/deacetylation. Data are the averages of two different points. “1×” corresponds to approximately 50 ng of HDAC1/2 in the complex. In panels A, B, and D, reaction mixtures were analyzed by SDS-PAGE, followed by Western blotting.