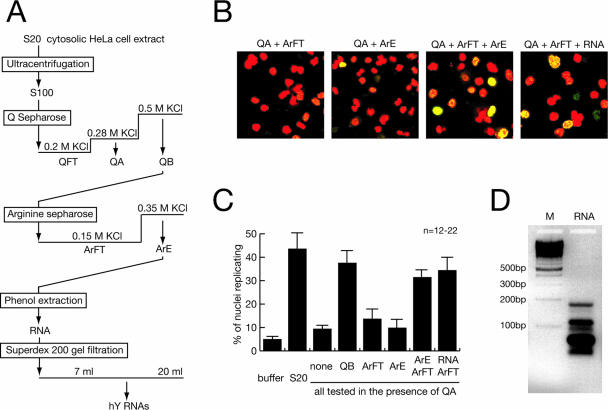
FIG. 1.
Purification of RNA as a factor for human chromosomal DNA replication. (A) Schematic diagram of fractionation steps. (B) Representative fields of nuclei replicating in vitro. Template nuclei from late-G1-phase cells were incubated with combinations of the indicated fractions, and replicating nuclei were detected by confocal fluorescence microscopy as detailed in references 36 and 37. Nuclear DNA is visualized by propidium iodide (red signal). Replicated DNA is labeled by incorporation of digoxigenin-dUMP, which is detected by fluorescein-conjugated antidigoxigenin Fab fragments (green signal). A merged signal appears in yellow. (C) Quantitative analysis of replicating G1-phase nuclei in vitro. Mean values and standard deviations of the proportions of replicating nuclei from the indicated reactions of 12 to 22 independent experiments (n) are shown (see Materials and Methods). Protein amounts per experiment were 100 μg unfractionated S20 cytosolic extract, 15 μg QA, 35 μg QB, 8 μg ArFT, a 20-μl volume of concentrated ArE containing less than 0.1 μg protein or the equivalent RNA prepared from this volume as specified. Fractions were used as indicated. (D) Visualization of RNA purified from fraction ArE. The RNA present in fraction ArE was purified by phenol extraction and isopropanol precipitation, separated on a 2% neutral agarose gel, and visualized by staining with ethidium bromide (lane RNA). A ladder of multimeric 100-bp DNA fragments was used as a molecular weight marker (lane M). An inverted image of the fluorescent gel is shown.