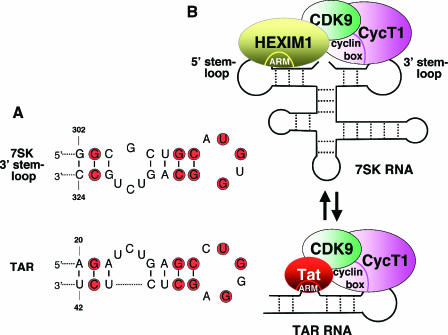
FIG. 4.
Architectural resemblance between the Tat-TAR-P-TEFb and HEXIM1-7SK-P-TEFb ribonucleoprotein complexes. (A) Comparison of nucleotide sequences between the G302-C324 apical region of the 3′ hairpin of 7SK RNA and the bulge-loop region of the HIV-1 TAR RNA (positions +20 to +42). The conserved nucleotides are shaded in red. Reprinted from reference 22 with permission.) (B) The Tat-TAR-P-TEFb and HEXIM1-7SK-P-TEFb complexes display many similar features and may share a common evolutionary origin. In addition to the sequence similarity between the HIV-1 TAR RNA and the 3′ hairpin of the 7SK RNA, both sequences also make direct contacts with CycT1. Moreover, both Tat and HEXIM1 directly interact with P-TEFb through binding to a region near the cyclin box in CycT1. Lastly, both HEXIM1 and Tat utilize a highly homologous and functionally interchangeable arginine-rich motif (ARM) for binding to their respective RNA partners. Given the multiple similarities shared between the two P-TEFb-containing ribonucleoprotein complexes, Tat has been shown to efficiently compete with HEXIM1 for binding to the same P-TEFb molecule. Tat can also disrupt the 7SK snRNP in an effort to convert the inactive P-TEFb into the active Tat/TAR-bound form for stimulating HIV-1 transcription. The secondary structure of 7SK is a modified version based on two previous reports (77, 92). For simplicity, only one copy each of HEXIM1, CDK9, and CycT1 is shown, although two copies of each are believed to interact with one 7SK molecule to form one HEXIM1-7SK-P-TEFb snRNP.