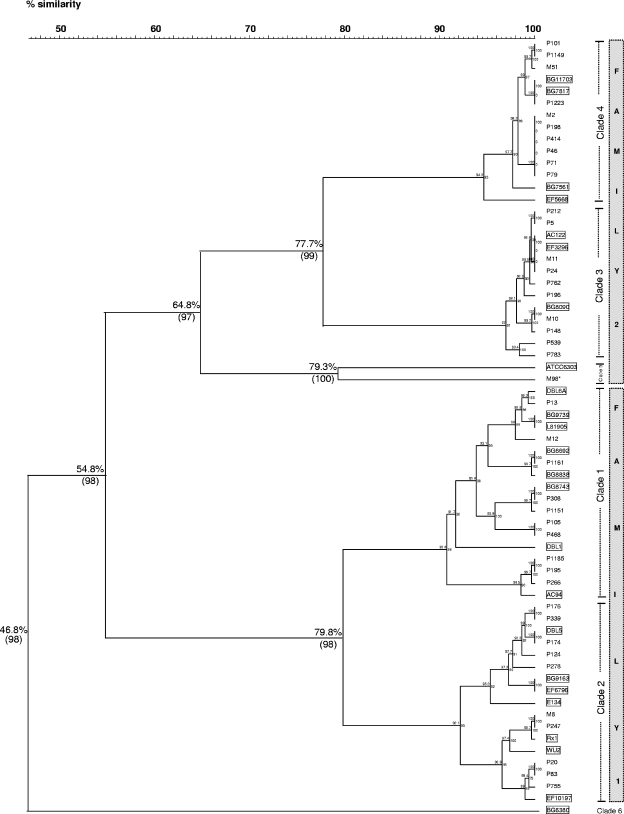
FIG. 2.
Dendrogram derived from comparison of 40 PspA-deduced amino acid sequences of nasopharyngeal strains and 24 sequences of PspA invasive reference strains (GenBank accession numbers AF071802 to AF071818, AF071820, AF071821, AF071823, AF071824, AF071826, U89711, and M74122). The dendrogram was constructed by using BioNumerics software (v 4.0). The names of the NP strains are indicated to the right of each branch point of the tree. Strain M98, marked with an asterisk, represents an invasive pneumococcal isolate from the blood of a child with meningitis. The invasive reference strains are boxed. Pearson's coefficient and the UPGMA clustering method were used for the analysis. Bootstrap values based on 1,000 replicates (in parentheses) and the percentages of similarity are distinguished for important nodes. The PspA families are represented by the gray column next to the corresponding clades.