Abstract
Extraintestinal pathogenic (ExPEC) Escherichia coli strains of serotype O18:K1:H7 are mainly responsible for neonatal meningitis and sepsis in humans and belong to a limited number of closely related clones. The same serotype is also frequently isolated from the extraintestinal lesions of colibacillosis in poultry, but it is not well known to what extent human and avian strains of this particular serotype are related. Twenty-two ExPEC isolates of human origin and 33 isolates of avian origin were compared on the basis of their virulence determinants, lethality for chicks, pulsed-field gel electrophoresis (PFGE) patterns, and classification in the main phylogenetic groups. Both avian and human isolates were lethal for chicks and harbored similar virulence genotypes. A major virulence pattern, identified in 75% of the isolates, was characterized by the presence of F1 variant fimbriae; S fimbriae; IbeA; the aerobactin system; and genomic fragments A9, A12, D1, D7, D10, and D11 and by the absence of P fimbriae, F1C fimbriae, Afa adhesin, and CNF1. All but one of the avian and human isolates also belonged to major phylogenetic group B2. However, various subclonal populations could be distinguished by PFGE in relation to animal species and geographical origin. These results demonstrate that very closely related clones can be recovered from extraintestinal infections in humans and chickens and suggest that avian pathogenic E. coli isolates of serotype O18:K1:H7 are potential human pathogens.
Extraintestinal pathogenic Escherichia coli (ExPEC) strains may cause various infections in humans and animals. These strains are implicated in a large range of extraintestinal infections in humans, such as neonatal meningitis, septicemia, urinary tract infections, and pneumonia, as well as in animals, such as urinary tract infections in cats and dogs, septicemia in calves and pigs, and systemic colibacillosis in birds (28, 41, 52, 53). ExPEC strains are characterized by virulence factors that may be present in various combinations. They include adhesins (F1, P, and S fimbriae), iron-sequestering systems (aerobactin and iro gene clusters), hemolysin, capsules (K1, K5), and various factors whose functions are not yet completely elucidated (Tsh, IbeA, CNF1, CDT, TraT), (25, 28, 41, 54, 67). These virulence factors are preferentially present in particular genetic backgrounds, and it has been shown that ExPEC strains mainly belong to the phylogenetic group ECOR B2 and, to a lesser extent, to group D (20, 48).
Avian pathogenic E. coli (APEC) strains are responsible for avian colibacillosis, a systemic infection in poultry that starts in the respiratory tract (4, 16, 34). APEC strains show similarities with human ExPEC strains. Even though no specific set of virulence factors can be associated with APEC strains, most of the virulence genes that they possess are similar to those identified in human ExPEC strains (13, 16, 21, 33, 39, 42, 50, 52, 63). Various genomic islands have also been identified to be possibly involved in virulence (11, 18, 36, 47, 58). Furthermore, several studies have demonstrated that some APEC strains could belong to the same clones as human ExPEC strains (1, 70, 71).
Among ExPEC strains, O18 strains are frequently isolated from neonatal meningitis and septicemia (2, 9, 27, 32). They belong to a limited number of related clones and possess common virulence factors, among which the K1 capsule plays a major role in circumventing host defenses (2, 5, 25, 27, 44, 60). The O18:K1 serotype may also be isolated from avian colibacillosis (6, 7, 51, 63); and the virulence factor IbeA, which is involved in human neonatal meningitis, has been shown to be preferentially associated with O18:K1 APEC strains (21). Even though no extensive study has been devoted to the comparison of O18 APEC strains with human O18 ExPEC strains, the genetic properties of O18:K1 strains of avian origin allow the hypothesis that they could represent a source for human contamination.
In order to assess this hypothesis, we compared O18:K1 ExPEC strains of human and avian origin on the basis of the presence of virulence determinants and pulsed-field gel electrophoresis (PFGE) patterns.
MATERIALS AND METHODS
Bacterial strains.
A total of 55 extraintestinal clinical isolates of E. coli O18 were used in this study: 22 strains of human origin and 33 strains of avian origin. All strains had been isolated between 1989 and 2001 from various European countries and, with the exception of strains BEN2782 and BEN2783, were epidemiologically unrelated; strains BEN2782 and BEN2783 were isolated from the same patient on different dates.
The human isolates included 15 strains isolated in Spain from the blood of septicemic adult patients 38 to 90 years old and identified as O18:H7 (this study) and 7 strains from neonatal meningitis cases in The Netherlands previously serotyped as O18:K1:H7 and named SP1, SP3, SP15, SP28, SP38, SP44, and SP58, according to their references in the work of Johnson et al. (27). The 33 avian isolates had been recovered from the heart blood or liver of chickens or turkeys with clinical signs of colibacillosis in Spain (24 isolates), France (6 isolates), and Belgium (3 isolates). They were selected on the basis of their O18:H7 serotype from a collection of 1,601 avian E. coli strains collected in Europe (63).
Furthermore, various E. coli strains were used as positive controls in PCR assays: strain BEN2908 (58) for genes neuC, fimA, fimH, and ibeA; strain BEN2908 or strain CFT073 (69) for genomic fragments A9, A12, D1, D7, D10, and D11; strain MT189 (17) for genes felA and papC; strains J96 (43), CFT073, and 536 (22) for alleles papGI, papGII, and papGIII, respectively; strain χ7122 (19) for tsh and strain KH576 (72) for iutA; strains 536 and CFT073 for sfaS and focG, respectively; strain BM2-1 (14) for cnfI; strain A30 (35) for afa; and strain E6468/62 (59) for cdt. Strains from the ECOR collection (45) were a kind gift from Thomas Whittam. They were used as controls for phylogenetic grouping: ECOR26 (B1 group), ECOR50 (D group), and ECOR62 (B2 group). E. coli strain MG1655 (8) was used as a negative control for virulence typing and as a positive control for phylogenetic ECOR group A. The E. coli strains were routinely grown in Luria-Bertani broth at 37°C with aeration and stored at −70°C in 20% glycerol until they were used.
Serotyping.
The determination of O and H antigens was carried out by agglutination by the method described by Blanco et al. (6) and with all available O (O1 to O181) and H (H1 to H56) antisera. All antisera were obtained and absorbed with the corresponding cross-reacting antigens to remove the nonspecific agglutinins. The O antisera were produced in the Laboratorio de Referencia de Escherichia coli (Lugo, Spain), and the H antisera were obtained from the Statens Serum Institut (Copenhagen, Denmark).
The presence of the capsular antigen K1 was detected by amplification of the neuC gene, as described below, and was checked phenotypically with a monoclonal antibody by using the Wellcogen Neisseria meningitidis B/E. coli K1 kit (Oxoid).
ECOR grouping.
The strains were classified into the four main phylogenetic groups of the ECOR collection by PCR, as described by Clermont et al. (12), by using three primers pairs: chuA1 and chuA2, yjaA1 and yjaA2, and tspE4C2.1 and tspE4C2.2 (Table 1). Classification into phylogenetic ECOR groups A, B1, B2, and D was based on the amplification of the genes chuA and yjaA and of fragment tspE4C2.1, as described by Clermont et al. (12).
TABLE 1.
Primers used for PCR amplifications
| Gene | Primers | Primer sequence (5′-3′) | Size of PCR product (bp) | Annealing temp (°C) | Reference |
|---|---|---|---|---|---|
| fimAJ96 | fimA1 | CGGCTCTGTCCCTSAGT | 500 | 52 | This study |
| fimA2 | GTCGCATCCGCATTAGC | ||||
| fimAvMT78 | fimA201 | TCTGGCTGATACTACACC | 266 | 52 | 37 |
| fimA215 | ACTTTAGGATGAGTACTG | ||||
| fimH | fimH2 | GATCTTTCGACGCAAATC | 389 | 52 | 3 |
| fimH17 | CGAGCAGAAACATCGCAG | ||||
| neuC | neu1 | AGGTGAAAAGCCTGGTAGTGTG | 676 | 61 | This study |
| neu2 | GGTGGTACATCCCGGGATGTC | ||||
| felA | fel1 | GGTCAASCAGCTAAAAACGGTAAGG | 239 | 61 | This study |
| fel2 | CCTTCAGAAACAGTACCGCCATTCG | ||||
| papC | pap1 | GACGGCTGTACTGCAGGGTGTGGCG | 328 | 61 | 35 |
| pap2 | ATATCCTTTCTGCAGGGATGCAATA | ||||
| papGI | papGIF | TCGTGCTCAGGTCCGGAATTT | 461 | 63 | 40 |
| papGIR | TGGCATCCCCCAACATTATCG | ||||
| papGII | papGIIF | GGGATGAGCGGGCCTTTGAT | 490 | 63 | 24 |
| papGIIR | CGGGCCCCCAAGTAACTCG | ||||
| papGIII | papGIIIF | GGCCTGCAATGGATTTACCTGG | 258 | 63 | 24 |
| papGIIIR | CCACCAAATGACCATGCCAGAC | ||||
| sfaS | sfaSF | GTGGATACGACGATTACTGTG | 242 | 63 | 29 |
| sfaSR | CCGCCAGCATTCCCTGTATTC | ||||
| focG | focGF | CAGCACAGGCAGTGGATACGA | 362 | 63 | 29 |
| focGR | GAATGTCGCCTGCCCATTGCT | ||||
| afa | afa1 | GCTGGGCAGCAAACTGATAACTCTC | 750 | 65 | 7, 35 |
| afa2 | CATCAAGCTGTTTGTTCGTCCGCCG | ||||
| iutA | iutA1 | ATGAGCATATCTCCGGACG | 587 | 55 | This study |
| iutA15 | CAGGTCGAAGAACATCTGG | ||||
| tsh | tsh03 | GGTGGTGCACTGGAGTGG | 640 | 55 | 19 |
| tsh15 | AGTCCAGCGTGATAGTGG | ||||
| ibeA | ibeAF | TGAACGTTTCGGTTGTTTTG | 814 | 55 | 21 |
| ibeAR | TGTTCAAATCCTGGCTGGAA | ||||
| cdt | cdt-s1 | GAAAGTAAATGGAATATAAATGTCCG | 466 | 55 | 65 |
| cdt-s2 | GAAAATAAATGGAACACACATGTCCG | ||||
| cdt-as1 | AAATCACCAAGAATCATCCAGTTA | 466 | 55 | 65 | |
| cdt-as2 | AAATCTCCTGCAATCATCCAGTTA | ||||
| cnfI | cnf-1S | GGGGGAAGTACAGAAGAATTA | 1,111 | 55 | 65 |
| cnf-1R | TTGCCGTCCACTCTCACCAGT | ||||
| A9 | 13F | TTTCGACTGCTGGATGAAC | 934 | 50 | 58 |
| 13R | AATCATGATTGACCGTGC | ||||
| A12 | 16F | ATGCACTCGATAAAAAAAGT | 860 | 50 | 58 |
| 16R | TTAAGAAGGTCGATATACGT | ||||
| D1 | 17F | ATGAATTCACAATTACTGGC | 1,998 | 50 | 58 |
| 17R | TTAGCTGTTCAGTAGCTCAC | ||||
| D7 | cat31 | TCAGTAAGAACGAAAGTGTG | 565 | 50 | This study |
| cat32 | ACAGGAACAATCCCGTGGAT | ||||
| D10 | D10F2 | ATCTTTACCGTCCTCACC | 135 | 50 | This study |
| D10R2 | CGTACCGCCTTCATTATC | ||||
| D11 | 20F | ATGCTGAACATGCAACAACA | 1,230 | 50 | 58 |
| 20R | TCAACCCTGTAGTAAACCAAT | ||||
| chuA | chuA.1 | GACGAACCAACGGTCAGGAT | 279 | 59 | 12 |
| chuA.2 | TGCCGCCAGTACCAAAGACA | ||||
| yjaA | yjaA.1 | TGAAGTGTCAGGAGACGCTG | 211 | 59 | 12 |
| yjaA.2 | ATGGAGAATGCGTTCCTCAAC | ||||
| tspE4 | tspE4C2.1 | GAGTAATGTCGGGGCATTCA | 152 | 59 | 12 |
| tspE4C2.2 | CGCGCCAACAAAGTATTACG |
Detection of cytotoxic activities of bacterial lysates and supernatants.
Experiments and the preparation of bacterial lysates were conducted as described previously (65). Briefly, nonconfluent HeLa cell monolayers in 96-well plates were infected with culture supernatants and with sonic lysates obtained from 48-h bacterial cultures. The plates were incubated at 37°C in a 5% CO2 atmosphere for 4 days. Morphological changes in the HeLa cells characteristic of the activity of cytolethal distending toxin were determined. E. coli strain DH5α (56) was used as a negative control, and all experiments were conducted in triplicate.
Virulence for chicks.
The virulence of each E. coli isolate was determined by using a test of lethality for 1-day-old chicks, as described previously (15). Briefly, groups of five 1-day-old specific-pathogen-free chickens were inoculated subcutaneously with a 24 h Luria-Bertani broth culture (about 108 CFU), and the mortality was recorded 4 days postinoculation.
Virulence genotyping.
Virulence genes were detected by PCR amplification carried out in a Perkin-Elmer temperature cycler 9700 (Applied Biosystems). The primers used are listed in Table 1. DNA crude extracts were prepared by a rapid boiling method. Six multiplex PCR assays were designed to detect simultaneously (i) fimA, the fimA variant (fimAvMT78), and fimH; (ii) neuC, felA, and papC; (iii) papGI, papGII, and papGIII; (iv) sfaS and focG; (v) tsh and iutA; and (vi) cdt. The other genes were detected in simple PCR assays. DNA fragments were amplified in a 25-μl PCR mixture with 1 U of Taq DNA polymerase (Promega), 25 pmol of the forward and reverse primers, and 5 nmol of each deoxynucleoside triphosphate (Promega) in 1× buffer. PCR conditions were as follows: 94°C for 3 min, followed by 30 cycles of 94°C for 1 min, the annealing temperature for 1 min, and 72°C for at least 30 s, according to the size of the amplified fragment (1 kbp/min), and then a final extension at 72°C for 10 min. Data were statistically analyzed by the chi-square test.
PFGE.
PFGE was conducted as described previously (58). Briefly, agarose plugs were prepared from a bacterial culture grown in brain heart infusion broth to an optical density at 600 nm of 1.0. Following incubation in a lysozyme solution (10 mM Tris HCl, pH 9, 100 mM EDTA, 5 mg ml−1 lysozyme, 0.05% Sarkosyl), they were then incubated overnight at 55°C (without shaking) in a lysis solution (10 mM Tris HCl, pH 9, 100 mM EDTA, 1 mg ml−1 proteinase K, 1% sodium dodecyl sulfate) and washed three times for 1 h each time in TE buffer (10 mM Tris-HCl, pH 8, 1 mM EDTA). After equilibration in the appropriate restriction enzyme incubation buffer, half plugs were digested overnight with 10 units of the enzyme XbaI (Takara Bio Europe). Pulsed-field gel electrophoresis was conducted in a CHEF-DRIII apparatus (Bio-Rad). The gels (1% agarose) were run at 14°C for 24 h in TBE buffer (Tris, 4 mM; borate, 4 mM; EDTA, 1 mM; pH 8.3) at 6 V cm−1. The pulse times were increased from 10 to 30 s. A bacteriophage lambda ladder CHEF DNA size standard (Bio-Rad) was used as a molecular size marker. The PFGE profiles of the strains were compared by using the Pearson similarity coefficient. The resulting dendrogram was calculated by using the unweighted pair group method with arithmetic averages (UPGMA) (62) included in the GelCompar software (Applied-Maths, St-Martens-Latem, Belgium).
PLS.
Partial least-squares (PLS) regression was also used, as described previously (38), to investigate the relationships between the PFGE profiles (variables X) and the classification of isolates as a function of their origins (avian strains versus human strains and strains from Spain versus strains from other countries for avian strains) (variables Y). PLS regression was established with SIMCA-P software, version 9.0 (UMETRI, Umea, Sweden).
The principle of this analysis is to search for regions of the PFGE profiles that could explain the origins of the strains (animal versus human or Spain versus other countries). Briefly, PLS components t[1], t[2],… , t[n], which are linear combinations of variables X, are calculated as follows: t[1] = w*11X1 + w*12X2 + … + w*1nXn, t[2] = w*21X1 + w*22X2 + … + w*2nXn,… , t[n] = w*n1X1 + w*n2X2 + … + w*nnXn. They describe the variables X and explain the variables Y. The w* terms denote the X loadings. The number of useful PLS components is determined by cross-validation (61): some of the PFGE profiles are not used for model development, and their values are then predicted by the model and compared with the actual values. The prediction error sum of squares (PRESS) is the squared differences between the observed and the predicted values for the data kept out of the model-fitting process. This procedure is repeated several times until each PFGE profile has been kept out once and only once. The final PRESS then has contributions from all data. For each component, SIMCA computes the overall PRESS/SS, where SS is the residual sum of squares of the previous component. A component is considered significant if PRESS/SS is statistically smaller than 1.0. Then, regressions relating the variables Y to the PLS components t[1], t[2],… , t[n] are built as follows: Y1 = c11t[1] + c12t[2] + … + c1nt[n] + residuals, Y2 = c21t[1] + c22t[2] + … + c2nt[n] + residuals,… , Yn = cn1t[1] + cn2t[2] + … + cnnt[n] + residuals. The c terms denote the Y loadings. The results of the analysis are visualized as groups of strains located in a plane defined by the PLS components. The explanatory performance of the models (expressed in percent) is evaluated by using the R2Y coefficient, which corresponds to the part of the variation of the Y matrix, as explained by the PLS components.
RESULTS
Avian and human isolates harbored similar virulence genotypes.
It was confirmed that all 55 strains expressed the O18 and the H7 antigens. All but one E. coli strain (human isolate BEN2794) possessed the neuC gene and also expressed the K1 capsular antigen. PCR was used to search for the presence of various virulence genes and of genomic fragments previously identified in APEC strain BEN2908 to be putatively associated with the virulence of ExPEC isolates (58).
The prevalence of virulence genes and genomic fragments was similar in both populations of human and avian isolates, and no statistically significant difference in their prevalence could be shown by the chi-square test (Table 2).
TABLE 2.
Prevalence of virulence genes and of phylogenetic groups in avian and human ExPEC isolates
| Gene, sequence, or group | % Positive
|
|
|---|---|---|
| Human isolates (n = 22) | Avian isolates (n = 33) | |
| Genes | ||
| neuC | 95.5 | 100 |
| fimAMG1655 | 0 | 0 |
| fimAvMT78 | 100 | 100 |
| fimH | 100 | 100 |
| felA | 0 | 0 |
| papC | 0 | 0 |
| papG | 0 | 3a |
| sfaS | 100 | 97 |
| focG | 0 | 0 |
| afa | 0 | 3 |
| iutA | 86.3 | 84.8 |
| ibeA | 95.5 | 100 |
| tsh | 45.5 | 42.4 |
| cdt | 63.6 | 63.6 |
| cnf1 | 4.5 | 3 |
| Sequences | ||
| A9 | 100 | 84.8 |
| A12 (sitA) | 100 | 97 |
| D1 | 100 | 93.9 |
| D7 | 100 | 97 |
| D10 | 100 | 100 |
| D11 (iroD) | 100 | 97 |
| Phylogenetic groups | ||
| ECOR B2 | 95.5 | 100 |
| ECOR B1 | 4.5 | 0 |
papGII.
More than 97% of strains in each group possessed genes of the fim operon and the sfa operon, but genes encoding other adhesins (P fimbriae, F1C fimbriae, or Afa) could not be detected. Only one avian strain (strain BEN806) possessed the papGII gene.
Interestingly, all isolates harbored a particular allele of fimA (fimAvMT78) that had previously been described in APEC strains (37, 66). The sequence of this fimA variant differs from the fimA sequence in strains MG1655 (K-12) and J96 by six variable domains (31, 46). It was detected by using the specific primers fimA201 and fimA215 (Table 1), whose sequences correspond to those of the first variable domain (amino acids 24 to 28, the characteristic N-terminal region of mature FimA) and the fourth variable domain (amino acids 104 to 109) of the FimA precursor, respectively. The complete sequences of the fimA genes of APEC strains BEN79 and BEN374 were obtained (data not shown), and it was confirmed that they were identical to that of fimAvMT78 (GenBank accession no. Z37500).
Gene ibeA, which is involved in penetration across the blood-brain barrier, was present in all but one strain (human strain SP1).
The aerobactin system was also present in more than 85% of the strains, as shown by the amplification of the iutA gene, which encodes the aerobactin receptor.
All sequences corresponding to genomic fragments identified in APEC strain BEN2908 following a subtractive hybridization (58) showed a high prevalence in avian isolates (85%) and were even present in 100% of isolates of human origin.
Genes tsh (which codes for an autotransporter) and cdt (which codes for a cytolethal distending toxin) showed a significantly lower prevalence compared with the prevalences of the other genes, and the cnf1 gene could be detected in only one avian isolate (BEN79) and one human isolate (BEN2794).
Thus, the major virulence pattern of avian and human ExPEC isolates of serotype O18:K1:H7 could be characterized by the presence of F1 variant fimbriae; S fimbriae; IbeA; the aerobactin system; and genomic fragments A9, A12, D1, D7, D10, and D11, as well as by the absence of P fimbriae, F1C fimbriae, the Afa adhesin, and CNF1. This pattern was present in 75% of the strains studied.
Two virulence genes, tsh and cdt, were not regularly present in human strains or in avian strains. The presence of the cdt gene was perfectly correlated with the expression of the cytolethal distending toxin, as demonstrated by the cytotoxic effect on HeLa cells (data not shown).
Both avian and human isolates were virulent for chicks.
All human isolates were lethal for 1-day-old chicks (at least one of five chicks inoculated was dead), and most of them (77.2%) killed five of five inoculated chicks. Among the avian strains, 72.7% of isolates killed five of five inoculated chicks, but four isolates (12%) did not kill any chicks. The last result was checked in a second test.
Two control strains were inoculated in the same experiment: virulent APEC strain MT512 (58), which killed five of five chicks, and avirulent strain EC79 (58), which did not kill any chicks.
Both avian and human isolates belonged to the same phylogenetic major ECOR group, group B2, but various subclonal populations were discriminated by PFGE.
As determined by the PCR assay of Clermont et al. (12), all but one strain (BEN2779) were assigned to phylogenetic ECOR group B2 (Table 2), which includes a majority of strains involved in extraintestinal infections (48).
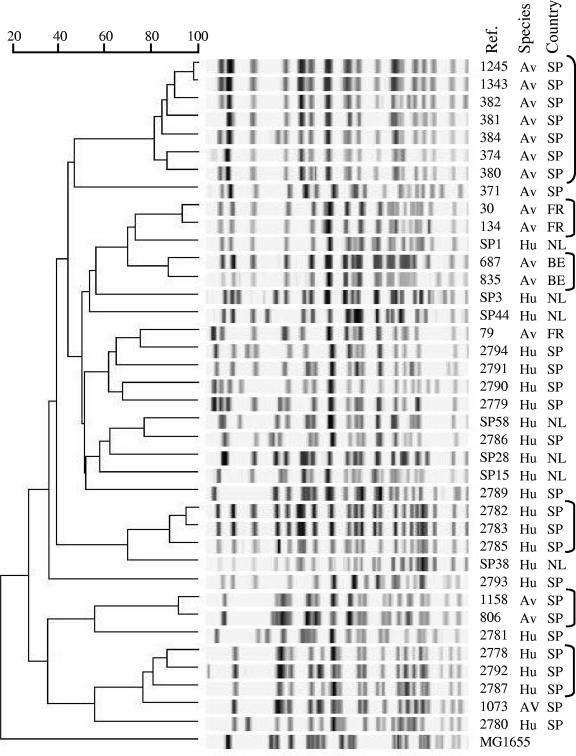
In order to get a better discrimination of the strains, we compared the PFGE profiles of the 22 ExPEC strains of human origin and the 16 avian ExPEC strains. The 38 strains studied showed 37 different electrophoretic patterns (strains BEN2782 and BEN2783, isolated from the same patient, showed identical PFGE profiles), and their similarities were calculated. The dendrogram obtained (Fig. 1) showed a group of seven APEC strains (strains BEN1245, BEN1343, BEN382, BEN381, BEN384, BEN374, and BEN380) with more than 80% similarity. All these strains had been isolated from diseased chickens in Spain. Other avian strains showing more than 80% similarity were BEN30 and BEN134 (isolated from turkeys in France), BEN687 and BEN835 (isolated from chickens in Belgium), and BEN806 and BEN1158 (isolated from chickens in Spain). Moreover, two groups of human ExPEC strains with similarities higher than 80% were identified: strains BEN2782, BEN2783, and BEN2785 and strains BEN2778, BEN2792, and BEN2787. All these strains had been isolated in Spain. No relationships with higher than 80% similarity were observed between avian and human ExPEC strains.
FIG. 1.
Genetic relationships among 38 ExPEC isolates of human and avian origin. The PFGE profiles obtained by XbaI restriction were compared by using the Pearson similarity coefficient, and the resulting dendrogram was calculated by the UPGMA method. The origins of the isolates are indicated: Av, avian; Hu, human; FR, France; SP, Spain; NL, The Netherlands. Groups of strains showing the highest similarity are indicated with brackets.
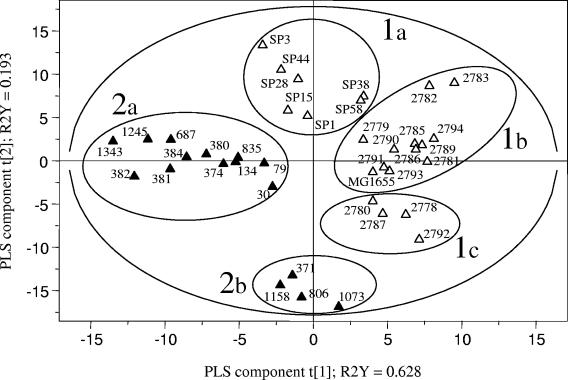
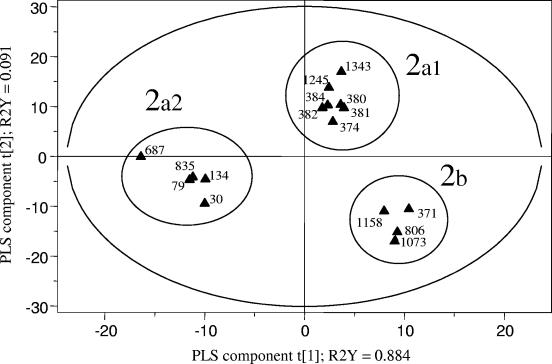
A PLS regression analysis based on the PFGE profiles confirmed that the avian ExPEC strains could be clearly differentiated from the human ExPEC strains (including nonpathogenic strain MG1655) in the 95% probability region (Fig. 2). However, these major groups were not homogeneous; and among the human strains three subgroups, groups 1a, 1b, and 1c (Fig. 2), that were related by both the geographical origin of the strains and the presence and expression of the cdt gene could be differentiated. No other correlation with virulence genes could be detected. All seven strains belonging to group 1a were from The Netherlands, and all but one (strain SP15) were cdt positive. The four strains in group 1c were from Spain and were cdt negative. All 11 strains in group 1b (except for strain MG1655) were also from Spain but showed heterogeneity for the presence of the cdt gene. The PLS regression analysis also revealed the heterogeneity of the avian ExPEC strains according to the presence and the expression of the cdt gene. In Fig. 2, groups 2a and 2b include cdt-positive and cdt-negative avian ExPEC strains, respectively. Moreover, a PLS regression analysis conducted with the avian ExPEC strains only (Fig. 3) showed that group 2a was not homogeneous, and cdt-positive APEC strains could be discriminated, according to their geographical origins, into subgroup 2a1 (seven strains from Spain) and subgroup 2a2 (five strains from France and Belgium). Group 2b was homogeneous and included only four cdt-negative APEC strains from Spain.
FIG. 2.
PLS discrimination between PFGE profiles of E. coli strains of human origin (open triangles; n = 23) and of avian origin (black triangles; n = 16). The R2Y coefficients correspond to the part of the variation of the Y matrix, as explained by the PLS components, and the explanatory performance of the model is evaluated by adding the R2Y coefficients. The cross-validation led to three PLS components (only the results for t[1] and t[2] are represented here), and the corresponding PLS model explained 91.6% of the variation of the Y matrix. The 95% probability region defined by the model is delimited by the ellipse. Five groups of strains could be distinguished and are delimited by circles: 1a, human cdt-positive strains (except SP15); 1b, human cdt-positive and cdt-negative strains; 1c, human cdt-negative strains; 2a, avian cdt-positive strains; and 2b, avian cdt-negative strains.
FIG. 3.
PLS discrimination between PFGE profiles of avian E. coli strains of different geographical origins. The R2Y coefficients correspond to the part of the variation of the Y matrix, as explained by the PLS components, and the explanatory performance of the model is evaluated by adding the R2Y coefficients. The cross-validation led to two PLS components, represented here as t[1] and t[2]. The corresponding PLS model explained 97.5% of the variation of the Y matrix. The 95% probability region defined by the model is delimited by the ellipse. The 2a group of cdt-positive strains could be subdivided into two subgroups, according to the geographical origins of the strain: 2a1, avian cdt-positive strains from Spain, and 2a2, cdt-positive strains from France and Belgium. The 2b group comprised cdt-negative strains from Spain only.
DISCUSSION
As shown by previous studies, some APEC strains are closely related to a recognized widespread clone that includes human ExPEC strains isolated from meningitis and septicemia (2, 71). Most of these studies included strains of various serogroups; thus, it is not clear to what extent APEC and human strains are identical and whether APEC strains could represent a zoonotic risk. In order to answer these questions more accurately, we focused our work on a collection of avian and human ExPEC strains belonging to the same serotype, serotype O18:K1:H7, which is one of the predominant serotypes both in neonatal septicemia and meningitis and in avian colibacillosis.
Our results confirm previous findings showing that ExPEC strains of human and avian origin share several virulence factors (26, 50). However, as pointed out by Mokady et al., only a few common virulence factors are present in nearly all the strains and thus can be considered important or even essential for the infectious process (41).
In the present study no statistically significant difference was observed in the prevalence of the 22 virulence genes and genomic fragments searched for in the human and the avian strains tested. Furthermore, 75% of the strains tested harbored the same virulence genotype, characterized by fimAvMT78; fimH; neuC; sfaS; ibeA; iutA; and genomic fragments A9, A12, D1, D7, D10, and D11. With the exception of the fim genes, all these genes usually belong to genomic regions that are absent from the genome of E. coli MG1655.
The corresponding virulence factors have been clearly associated with the pathogenicities of ExPEC strains for humans and/or chickens, as demonstrated for K1 (30, 39), F1 fimbriae (64), S fimbriae (10, 57), IbeA (21, 23), and the aerobactin-sequestering system (10, 33). The translated sequences of genomic fragments A12 and D11 from APEC strain BEN2908 are both homologous with proteins SitA and IroD, respectively, which are involved in iron acquisition (58). These proteins are considered potential virulence factors in the human strain E. coli CFT073 and in APEC strains (18, 55, 69). The putative role in virulence of the other genomic fragments (A9, D7, and D10) is determined on the basis of their higher incidence in pathogenic avian E. coli strains than in nonpathogenic strains and on their presence in several human ExPEC strains (58).
It is noticeable that all strains tested possessed a particular variant of fimA that was first described in APEC strain MT78 (GenBank accession no. Z37500) and then in various APEC strains (37, 66). This allele is identical to the fimA allele of the archetypal strains RS218 and IHE3034 that was recently specified by Weissman et al. as the fimA C0 allele (68). The presence of this particular allele indicates phylogenetic relationship between the strains; indeed, it was shown to be present in K1 ExPEC strains of the phylogenetic ECOR group B2 belonging to the sequence type 95 complex (68).
The possession of a set of homologous virulence genes by avian and human strains suggests that they have similar virulence mechanisms. Moreover, all human strains were lethal when they were inoculated subcutaneously into 1-day-old chicks. However, when the physiopathologies of both infections are compared, the portals of entry of E. coli are different between chickens and human newborns. Avian colibacillosis initiates in the upper respiratory tract, following injury of the tracheal epithelium by viruses or mycoplasma, and then APEC cells colonize the air sacs and lungs and invade the bloodstream (4). In the case of neonatal meningitis, E. coli first colonizes the intestinal tract and then translocates from the intestinal lumen to the bloodstream (9, 49). As specific colonization factors have not yet been identified in human isolates or in APEC isolates, inoculation of a human strain into chickens via the intratracheal route could bring a first answer to the question of possible cross-contamination with O18:K1:H7 strains.
The results obtained here suggest strong genetic relationships between human ExPEC strains and APEC strains of serotype O18:K1:H7. All but one of the strains tested belonged to phylogenetic ECOR group B2 and had the classic meningitis-associated traits (sfa and ibeA). White et al. (71), comparing avian and human ExPEC strains by multilocus enzyme electrophoresis, showed that O18:H7 isolates of both origins were located in the same “cluster D” and were closely related to the O18:K1 clone originally identified by Achtman et al. (2). As demonstrated by Achtman et al. (2), the O18:K1 clone can be subdivided into two subclones that correspond to outer membrane protein patterns 6 and 9 (OMP6 and OMP9, respectively) and that are related to the geographical origins of the strains. In our study, the European origin of the isolates is consistent with their belonging to the OMP9 subclone. Moreover, the lack of the pap genes is another characteristic of OMP9 E. coli isolates, as pointed out by Johnson et al. (27).
Belonging to this widespread clone are also human strains SP1, SP3, SP15, SP28, SP38, SP44, and SP58, as demonstrated by Johnson et al. (27), who identified a large “cluster II” that comprised predominantly O18:K1 neonatal meningitis-associated isolates.
Thus, our results demonstrate that avian and human ExPEC strains of serotype O18:K1:H7 show strong similarities according to their virulence genotypes and phylogenetic groups. However, by using PFGE, which is considered a highly discriminating method, 37 different PFGE profiles were obtained; and cluster analysis by UPGMA revealed that only a few subclusters that had more than 80% similarity were observed, and the subclusters never comprised both human and avian isolates. PLS regression analysis of the PFGE profiles showed that the strains could be distinguished according to their human or avian origin. The five groups resulting from PLS regression analysis (Fig. 2) clearly showed that overlap between human and avian strains does not exist, thus demonstrating some diversity among isolates of clone O18:K1:H7. Moreover, strains were grouped according to their geographical origin (Spain, France and Belgium, or The Netherlands), showing the local dissemination of closely related clones. It is noticeable that groups defined by PLS regression analysis included cdt-positive isolates only or cdt-negative isolates only, with the exception of group 1b, which included both types of isolates. This observation is in favor of the parallel evolution of different clones with close genetic relationships.
Studying a collection of O18:K1:H7 E. coli isolates from women with acute cystitis, healthy control patients, and infants with neonatal meningitis, Johnson et al. (25) demonstrated a high degree of commonality between these strains on the basis of randomly amplified polymorphic DNA analysis, nicotinamide auxotrophy, outer membrane protein patterns, and virulence factor profiles. Our results show that even though O18:K1:H7 E. coli strains seem to be very closely related according to their virulence genotypes and their phylogenetic groups, they can be differentiated by a highly discriminating method such as PFGE. The results obtained with the set of strains that we have studied here show that various but closely related clones can be recovered from extraintestinal infections in humans and chickens. Epidemiological studies are required to demonstrate if they have evolved independently or if cross-contamination between human and avian communities is possible, leading to the hypothesis that avian colibacillosis due to O18:K1:H7 strains could be considered a zoonosis.
Acknowledgments
We thank María Pilar Alonso (Unidad de Microbioloxía, Complexo Hospitalario, Xeral-Calde, Lugo, Spain), Thomas Whittam, James Johnson, and Lodewijk Spanjaard for the kind gift of several strains used in this study. Thanks are also due to Olivier Marchès for the identification of cdt-positive strains. Laila Bakri and Katia Courvoisier are greatly acknowledged for their skillful technical assistance with the PCR experiments.
This work was funded in part by the European Community (contract FAIR6-CT-98-4093).
REFERENCES
- 1.Achtman, M., M. Heuzenroeder, B. Kusecek, H. Ochman, D. Caugant, R. K. Selander, V. Vaisanen-Rhen, T. K. Korhonen, S. Stuart, F. Orskov, et al. 1986. Clonal analysis of Escherichia coli O2:K1 isolated from diseased humans and animals. Infect. Immun. 51:268-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Achtman, M., A. Mercer, B. Kusecek, A. Pohl, M. Heuzenroeder, W. Aaronson, A. Sutton, and R. P. Silver. 1983. Six widespread bacterial clones among Escherichia coli K1 isolates. Infect. Immun. 39:315-335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arné, P., D. Marc, A. Brée, C. Schouler, and M. Dho-Moulin. 2000. Increased tracheal colonization in chickens without impairing pathogenic properties of avian pathogenic Escherichia coli MT78 with a fimH deletion. Avian Dis. 44:343-355. [PubMed] [Google Scholar]
- 4.Barnes, H. J., J.-P. Vaillancourt, and W. B. Gross. 2003. Colibacillosis, p. 631-652. In H. J. B. Y. M. Saif, J. R. Glisson, A. M. Fadly, L. R. McDougald, and D. E. Swayne (ed.), Diseases of poultry, 11th ed. Iowa State University Press, Ames.
- 5.Bingen, E., B. Picard, N. Brahimi, S. Mathy, P. Desjardins, J. Elion, and E. Denamur. 1998. Phylogenetic analysis of Escherichia coli strains causing neonatal meningitis suggests horizontal gene transfer from a predominant pool of highly virulent B2 group strains. J. Infect. Dis. 177:642-650. [DOI] [PubMed] [Google Scholar]
- 6.Blanco, J. E., M. Blanco, A. Mora, W. H. Jansen, V. Garcia, M. L. Vazquez, and J. Blanco. 1998. Serotypes of Escherichia coli isolated from septicaemic chickens in Galicia (northwest Spain). Vet. Microbiol. 61:229-235. [DOI] [PubMed] [Google Scholar]
- 7.Blanco, M., J. E. Blanco, M. P. Alonso, A. Mora, C. Balsalobre, F. Munoa, A. Juarez, and J. Blanco. 1997. Detection of pap, sfa, and afa adhesin-encoding operons in uropathogenic Escherichia coli strains: relationship with expression of adhesins and production of toxins. Res. Microbiol. 148:745-755. [DOI] [PubMed] [Google Scholar]
- 8.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 9.Bonacorsi, S., and E. Bingen. 2005. Molecular epidemiology of Escherichia coli causing neonatal meningitis. Int. J. Med. Microbiol. 295:373-381. [DOI] [PubMed] [Google Scholar]
- 10.Bonacorsi, S., O. Clermont, V. Houdouin, C. Cordevant, N. Brahimi, A. Marecat, C. Tinsley, X. Nassif, M. Lange, and E. Bingen. 2003. Molecular analysis and experimental virulence of French and North American Escherichia coli neonatal meningitis isolates: identification of a new virulent clone. J. Infect. Dis. 187:1895-1906. [DOI] [PubMed] [Google Scholar]
- 11.Chouikha, I., P. Germon, A. Brée, P. Gilot, M. Moulin-Schouleur, and C. Schouler. 2006. A selC-associated genomic island of the extraintestinal avian pathogenic Escherichia coli strain BEN2908 is involved in carbohydrate uptake and virulence. J. Bacteriol. 188:977-987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Clermont, O., S. Bonacorsi, and E. Bingen. 2000. Rapid and simple determination of the Escherichia coli phylogenetic group. Appl. Environ. Microbiol. 66:4555-4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delicato, E. R., B. G. de Brito, L. C. Gaziri, and M. C. Vidotto. 2003. Virulence-associated genes in Escherichia coli isolates from poultry with colibacillosis. Vet. Microbiol. 94:97-103. [DOI] [PubMed] [Google Scholar]
- 14.De Rycke, J., L. Phan-Thanh, and S. Bernard. 1989. Immunochemical identification and biological characterization of cytotoxic necrotizing factor from Escherichia coli. J. Clin. Microbiol. 27:983-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dho, M., and J. P. Lafont. 1984. Adhesive properties and iron uptake ability in Escherichia coli lethal and nonlethal for chicks. Avian Dis. 28:1016-1025. [PubMed] [Google Scholar]
- 16.Dho-Moulin, M., and J. M. Fairbrother. 1999. Avian pathogenic Escherichia coli (APEC). Vet. Res. 30:299-316. [PubMed] [Google Scholar]
- 17.Dho-Moulin, M., J. F. van den Bosch, J. P. Girardeau, A. Brée, T. Barat, and J. P. Lafont. 1990. Surface antigens from Escherichia coli O2 and O78 strains of avian origin. Infect. Immun. 58:740-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dozois, C. M., F. Daigle, and R. Curtiss III. 2003. Identification of pathogen-specific and conserved genes expressed in vivo by an avian pathogenic Escherichia coli strain. Proc. Natl. Acad. Sci. USA 100:247-252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dozois, C. M., M. Dho-Moulin, A. Brée, J. M. Fairbrother, C. Desautels, and R. Curtiss III. 2000. Relationship between the Tsh autotransporter and pathogenicity of avian Escherichia coli and localization and analysis of the Tsh genetic region. Infect. Immun. 68:4145-4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Escobar-Paramo, P., O. Clermont, A. B. Blanc-Potard, H. Bui, C. Le Bouguenec, and E. Denamur. 2004. A specific genetic background is required for acquisition and expression of virulence factors in Escherichia coli. Mol. Biol. Evol. 21:1085-1094. [DOI] [PubMed] [Google Scholar]
- 21.Germon, P., Y. H. Chen, L. He, J. E. Blanco, A. Brée, C. Schouler, S. H. Huang, and M. Moulin-Schouleur. 2005. ibeA, a virulence factor of avian pathogenic Escherichia coli. Microbiology 151:1179-1186. [DOI] [PubMed] [Google Scholar]
- 22.Hacker, J., S. Knapp, and W. Goebel. 1983. Spontaneous deletions and flanking regions of the chromosomally inherited hemolysin determinant of an Escherichia coli O6 strain. J. Bacteriol. 154:1145-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huang, S. H., C. Wass, Q. Fu, N. V. Prasadarao, M. Stins, and K. S. Kim. 1995. Escherichia coli invasion of brain microvascular endothelial cells in vitro and in vivo: molecular cloning and characterization of invasion gene ibe10. Infect. Immun. 63:4470-4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johnson, J. R., and J. J. Brown. 1996. A novel multiply primed polymerase chain reaction assay for identification of variant papG genes encoding the gal(alpha 1-4)gal-binding PapG adhesins of Escherichia coli. J. Infect. Dis. 173:920-926. [DOI] [PubMed] [Google Scholar]
- 25.Johnson, J. R., P. Delavari, and T. T. O'Bryan. 2001. Escherichia coli O18:K1:H7 isolates from patients with acute cystitis and neonatal meningitis exhibit common phylogenetic origins and virulence factor profiles. J. Infect. Dis. 183:425-434. [DOI] [PubMed] [Google Scholar]
- 26.Johnson, J. R., A. C. Murray, A. Gajewski, M. Sullivan, P. Snippes, M. A. Kuskowski, and K. E. Smith. 2003. Isolation and molecular characterization of nalidixic acid-resistant extraintestinal pathogenic Escherichia coli from retail chicken products. Antimicrob. Agents Chemother. 47:2161-2168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson, J. R., E. Oswald, T. T. O'Bryan, M. A. Kuskowski, and L. Spanjaard. 2002. Phylogenetic distribution of virulence-associated genes among Escherichia coli isolates associated with neonatal bacterial meningitis in The Netherlands. J. Infect. Dis. 185:774-784. [DOI] [PubMed] [Google Scholar]
- 28.Johnson, J. R., and T. A. Russo. 2002. Extraintestinal pathogenic Escherichia coli: “the other bad E. coli.” J. Lab. Clin. Med. 139:155-162. [DOI] [PubMed] [Google Scholar]
- 29.Johnson, J. R., and A. L. Stell. 2000. Extended virulence genotypes of Escherichia coli strains from patients with urosepsis in relation to phylogeny and host compromise. J. Infect. Dis. 181:261-272. [DOI] [PubMed] [Google Scholar]
- 30.Kim, K. J., S. J. Elliott, F. Di Cello, M. F. Stins, and K. S. Kim. 2003. The K1 capsule modulates trafficking of Escherichia coli-containing vacuoles and enhances intracellular bacterial survival in human brain microvascular endothelial cells. Cell. Microbiol. 5:245-252. [DOI] [PubMed] [Google Scholar]
- 31.Klemm, P. 1984. The fimA gene encoding the type-1 fimbrial subunit of Escherichia coli. Nucleotide sequence and primary structure of the protein. Eur. J. Biochem. 143:395-399. [DOI] [PubMed] [Google Scholar]
- 32.Korhonen, T. K., M. V. Valtonen, J. Parkkinen, V. Vaisanen-Rhen, J. Finne, F. Orskov, I. Orskov, S. B. Svenson, and P. H. Makela. 1985. Serotypes, hemolysin production, and receptor recognition of Escherichia coli strains associated with neonatal sepsis and meningitis. Infect. Immun. 48:486-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lafont, J. P., M. Dho, H. M. D'Hauteville, A. Brée, and P. J. Sansonetti. 1987. Presence and expression of aerobactin genes in virulent avian strains of Escherichia coli. Infect. Immun. 55:193-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.La Ragione, R. M., and M. J. Woodward. 2002. Virulence factors of Escherichia coli serotypes associated with avian colisepticaemia. Res. Vet. Sci. 73:27-35. [DOI] [PubMed] [Google Scholar]
- 35.Le Bouguénec, C., M. Archambaud, and A. Labigne. 1992. Rapid and specific detection of the pap, afa, and sfa adhesin-encoding operons in uropathogenic Escherichia coli strains by polymerase chain reaction. J. Clin. Microbiol. 30:1189-1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li, G., C. Laturnus, C. Ewers, and L. H. Wieler. 2005. Identification of genes required for avian Escherichia coli septicemia by signature-tagged mutagenesis. Infect. Immun. 73:2818-2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marc, D., and M. Dho-Moulin. 1996. Analysis of the fim cluster of an avian O2 strain of Escherichia coli: serogroup-specific sites within fimA and nucleotide sequence of fimI. J. Med. Microbiol. 44:444-452. [DOI] [PubMed] [Google Scholar]
- 38.Matte-Tailliez, O., E. Lepage, M. Tenenhaus, and P. Tailliez. 2002. Use of predictive modeling for Propionibacterium strain classification. Syst. Appl. Microbiol. 25:386-395. [DOI] [PubMed] [Google Scholar]
- 39.Mellata, M., M. Dho-Moulin, C. M. Dozois, R. Curtiss III, P. K. Brown, P. Arné, A. Brée, C. Desautels, and J. M. Fairbrother. 2003. Role of virulence factors in resistance of avian pathogenic Escherichia coli to serum and in pathogenicity. Infect. Immun. 71:536-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mitsumori, K., A. Terai, S. Yamamoto, and O. Yoshida. 1998. Identification of S, F1C and three PapG fimbrial adhesins in uropathogenic Escherichia coli by polymerase chain reaction. FEMS Immunol. Med. Microbiol. 21:261-268. [DOI] [PubMed] [Google Scholar]
- 41.Mokady, D., U. Gophna, and E. Z. Ron. 2005. Extensive gene diversity in septicemic Escherichia coli strains. J. Clin. Microbiol. 43:66-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ngeleka, M., L. Brereton, G. Brown, and J. M. Fairbrother. 2002. Pathotypes of avian Escherichia coli as related to tsh-, pap-, pil-, and iuc-DNA sequences, and antibiotic sensitivity of isolates from internal tissues and the cloacae of broilers. Avian Dis. 46:143-152. [DOI] [PubMed] [Google Scholar]
- 43.Normark, S., D. Lark, R. Hull, M. Norgren, M. Baga, P. O'Hanley, G. Schoolnik, and S. Falkow. 1983. Genetics of digalactoside-binding adhesin from a uropathogenic Escherichia coli strain. Infect. Immun. 41:942-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ochman, H., and R. K. Selander. 1984. Evidence for clonal population structure in Escherichia coli. Proc. Natl. Acad. Sci. USA 81:198-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ochman, H., and R. K. Selander. 1984. Standard reference strains of Escherichia coli from natural populations. J. Bacteriol. 157:690-693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Orndorff, P. E., and S. Falkow. 1984. Organization and expression of genes responsible for type 1 piliation in Escherichia coli. J. Bacteriol. 159:736-744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Parreira, V. R., and C. L. Gyles. 2003. A novel pathogenicity island integrated adjacent to the thrW tRNA gene of avian pathogenic Escherichia coli encodes a vacuolating autotransporter toxin. Infect. Immun. 71:5087-5096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Picard, B., J. S. Garcia, S. Gouriou, P. Duriez, N. Brahimi, E. Bingen, J. Elion, and E. Denamur. 1999. The link between phylogeny and virulence in Escherichia coli extraintestinal infection. Infect. Immun. 67:546-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pluschke, G., A. Mercer, B. Kusecek, A. Pohl, and M. Achtman. 1983. Induction of bacteremia in newborn rats by Escherichia coli K1 is correlated with only certain O (lipopolysaccharide) antigen types. Infect. Immun. 39:599-608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rodriguez-Siek, K. E., C. W. Giddings, C. Doetkott, T. J. Johnson, M. K. Fakhr, and L. K. Nolan. 2005. Comparison of Escherichia coli isolates implicated in human urinary tract infection and avian colibacillosis. Microbiology 151:2097-2110. [DOI] [PubMed] [Google Scholar]
- 51.Rodriguez-Siek, K. E., C. W. Giddings, C. Doetkott, T. J. Johnson, and L. K. Nolan. 2005. Characterizing the APEC pathotype. Vet. Res. 36:241-256. [DOI] [PubMed] [Google Scholar]
- 52.Ron, E. Z. 2006. Host specificity of septicemic Escherichia coli: human and avian pathogens. Curr. Opin. Microbiol. 9:28-32. [DOI] [PubMed] [Google Scholar]
- 53.Russo, T. A., and J. R. Johnson. 2000. Proposal for a new inclusive designation for extraintestinal pathogenic isolates of Escherichia coli: ExPEC. J. Infect. Dis. 181:1753-1754. [DOI] [PubMed] [Google Scholar]
- 54.Russo, T. A., C. D. McFadden, U. B. Carlino-MacDonald, J. M. Beanan, T. J. Barnard, and J. R. Johnson. 2002. IroN functions as a siderophore receptor and is a urovirulence factor in an extraintestinal pathogenic isolate of Escherichia coli. Infect. Immun. 70:7156-7160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sabri, M., S. Léveillé, and C. M. Dozois. 2006. A SitABC homologue from an avian pathogenic Escherichia coli mediates transport of iron and manganese and resistance to hydrogen peroxyde. Microbiology 152:745-758. [DOI] [PubMed] [Google Scholar]
- 56.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 57.Saren, A., R. Virkola, J. Hacker, and T. K. Korhonen. 1999. The cellular form of human fibronectin as an adhesion target for the S fimbriae of meningitis-associated Escherichia coli. Infect. Immun. 67:2671-2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schouler, C., F. Koffmann, C. Amory, S. Leroy-Setrin, and M. Moulin-Schouleur. 2004. Genomic subtraction for the identification of putative new virulence factors of an avian pathogenic Escherichia coli strain of O2 serogroup. Microbiology 150:2973-2984. [DOI] [PubMed] [Google Scholar]
- 59.Scott, D. A., and J. B. Kaper. 1994. Cloning and sequencing of the genes encoding Escherichia coli cytolethal distending toxin. Infect. Immun. 62:244-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Selander, R. K., T. K. Korhonen, V. Vaisanen-Rhen, P. H. Williams, P. E. Pattison, and D. A. Caugant. 1986. Genetic relationships and clonal structure of strains of Escherichia coli causing neonatal septicemia and meningitis. Infect. Immun. 52:213-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.SIMCA-P9.0. 2001. A new standard in multivariate data analysis, p. 122. In User's guide and tutorial. Umetrics, Umea, Sweden.
- 62.Sokal, R. R., and C. D. Michener. 1958. A statistical method for evaluating systematic relationships. Univ. Kans. Sci. Bull. 38:1409-1438. [Google Scholar]
- 63.Stordeur, P., D. Marlier, J. Blanco, E. Oswald, F. Biet, M. Dho-Moulin, and J. Mainil. 2002. Examination of Escherichia coli from poultry for selected adhesin genes important in disease caused by mammalian pathogenic E. coli. Vet. Microbiol. 84:231-241. [DOI] [PubMed] [Google Scholar]
- 64.Teng, C. H., M. Cai, S. Shin, Y. Xie, K. J. Kim, N. A. Khan, F. Di Cello, and K. S. Kim. 2005. Escherichia coli K1 RS218 interacts with human brain microvascular endothelial cells via type 1 fimbria bacteria in the fimbriated state. Infect. Immun. 73:2923-2931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Toth, I., F. Herault, L. Beutin, and E. Oswald. 2003. Production of cytolethal distending toxins by pathogenic Escherichia coli strains isolated from human and animal sources: establishment of the existence of a new cdt variant (type IV). J. Clin. Microbiol. 41:4285-4291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vandemaele, F., D. Vandekerchove, M. Vereecken, J. Derijcke, M. Dho-Moulin, and B. M. Goddeeris. 2003. Sequence analysis demonstrates the conservation of fimH and variability of fimA throughout avian pathogenic Escherichia coli (APEC). Vet. Res. 34:153-163. [DOI] [PubMed] [Google Scholar]
- 67.Watt, S., P. Lanotte, L. Mereghetti, M. Moulin-Schouleur, B. Picard, and R. Quentin. 2003. Escherichia coli strains from pregnant women and neonates: intraspecies genetic distribution and prevalence of virulence factors. J. Clin. Microbiol. 41:1929-1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Weissman, S. J., S. Chattopadhyay, P. Aprikian, M. Obata-Yasuoka, Y. Yarova-Yarovaya, A. Stapleton, W. Ba-Thein, D. Dykhuizen, J. R. Johnson, and E. V. Sokurenko. 2006. Clonal analysis reveals high rate of structural mutations in fimbrial adhesins of extraintestinal pathogenic Escherichia coli. Mol. Microbiol. 59:975-988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Welch, R. A., V. Burland, G. Plunkett III, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.White, D. G., M. Dho-Moulin, R. A. Wilson, and T. S. Whittam. 1993. Clonal relationships and variation in virulence among Escherichia coli strains of avian origin. Microb. Pathog. 14:399-409. [DOI] [PubMed] [Google Scholar]
- 71.White, D. G., R. A. Wilson, A. S. Gabriel, M. Saco, and T. S. Whittam. 1990. Genetic relationships among strains of avian Escherichia coli associated with swollen-head syndrome. Infect. Immun. 58:3613-3620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Williams, P. H. 1979. Novel iron uptake system specified by ColV plasmids: an important component in the virulence of invasive strains of Escherichia coli. Infect. Immun. 26:925-932. [DOI] [PMC free article] [PubMed] [Google Scholar]