FIG.2.
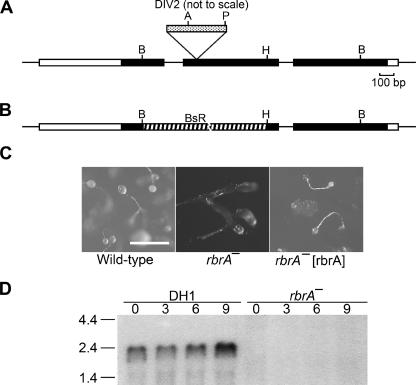
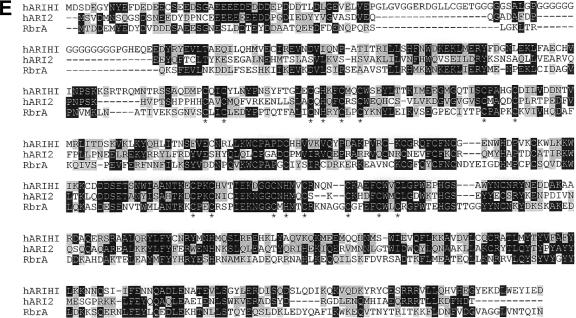
rbrA gene. (A) Genomic map and REMI-generated mutation. REMI was carried out by electroporating EcoRI-linearized DIV2 (the mutagenic plasmid carrying the Dictyostelium pyr5-6 gene) into DH1 (pyr5-6−) cells along with the restriction enzyme MunI and selecting for uracil prototrophs. The thick bar represents exons of rbrA, and the black portion of the thick bar designates the open reading frame. Restriction sites: A, Asp718; B, BglII; H, HindIII; P, PvuII. The location of the DIV2 insertion is identified (spotted bar; not drawn to scale). (B) Diagram of rbrA disrupted by blasticidin-resistance gene (striped bar) replacement. (C) Rescue of rbrA mutant development phenotype. Wild-type cells, rbrA− cells, and rbrA− cells carrying an rbrA expression plasmid (rbrA− [rbrA]) were grown on nutrient agar plates in association with Klebsiella aerogenes and then allowed to starve on the agar plates. Bar is 0.5 mm. (D) Expression of rbrA. RNA from the indicated cell lines developed for the indicated time in hours was resolved by electrophoresis through 1.2% agarose-formaldehyde gels, blotted onto nylon membranes, and hybridized with a probe specific for rbrA. Size standards are indicated on the left in kilobases. (E) Comparison of the predicted amino acid sequences of RbrA with human ARIHI and ARI2. The amino acid identities between RbrA (AAR10851; dictyBase DDB0191418), human ARIH1 (AAH51877), and human ARI2 (CAA10276) are in black, and amino acid similarities are in highlighted in gray. Conserved cysteine and histidine residues of the RING fingers are marked by an asterisk.