Abstract
The Hgt4 protein of Candida albicans (orf19.5962) is orthologous to the Snf3 and Rgt2 glucose sensors of Saccharomyces cerevisiae that govern sugar acquisition by regulating the expression of genes encoding hexose transporters. We found that HGT4 is required for glucose induction of the expression of HGT12, HXT10, and HGT7, which encode apparent hexose transporters in C. albicans. An hgt4Δ mutant is defective for growth on fermentable sugars, which is consistent with the idea that Hgt4 is a sensor of glucose and similar sugars. Hgt4 appears to be sensitive to glucose levels similar to those in human serum (∼5 mM). HGT4 expression is repressed by high levels of glucose, which is consistent with the idea that it encodes a high-affinity sugar sensor. Glucose sensing through Hgt4 affects the yeast-to-hyphal morphological switch of C. albicans cells: hgt4Δ mutants are hypofilamented, and a constitutively signaling form of Hgt4 confers hyperfilamentation of cells. The hgt4Δ mutant is less virulent than wild-type cells in a mouse model of disseminated candidiasis. These results suggest that Hgt4 is a high-affinity glucose sensor that contributes to the virulence of C. albicans.
Candida albicans is responsible for most yeast infections in humans. In addition to causing ordinary vaginal and oral (thrush) infections, this fungus is a serious threat to immunocompromised patients, for whom disseminated, invasive candidiasis is common and often fatal (31, 56). The fungus is normally maintained as a harmless commensal on skin, on mucosa, and in the gut (23) but becomes an opportunistic pathogen upon entry into the bloodstream of immunocompromised (particularly neutropenic) individuals (25, 45). Its “virulence” is due to its robust ability to access and thrive in various niches within its host, which provide an abundant source of microbes available to cause disease (1, 25, 26). Within the host, C. albicans must efficiently compete for nutrients with host cells and an extensive repertoire of resident microbes (3).
Glucose acts as a morphogen for C. albicans, triggering the yeast-to-hyphal transition that is a vital determinant of virulence (15). Mutant C. albicans strains that are “frozen” in either phase cause less mortality in mouse models of disseminated infection, underscoring the importance of morphological plasticity for virulence (47). The yeast form is thought to disseminate easily via body fluids, and the hyphal form can extravasate into tissues or form mycelial biofilms (6, 52).
Because glucose plays a central role as a carbon and energy source, glucose sensing and response is highly evolved and closely regulated in most organisms. The importance of glucose to yeast cells is illustrated by the large number of hexose transporters they possess: Saccharomyces cerevisiae has at least 17 different hexose transporters that are expressed under different conditions; C. albicans appears to have over 20 (11). S. cerevisiae is thought to require this large number of hexose transporters because it prefers to ferment glucose, a lifestyle that demands a high influx of glucose because it yields only two molecules of ATP for each molecule of glucose metabolized. It is somewhat surprising that C. albicans, which is thought to prefer respiration and therefore can garner up to 38 ATPs per glucose molecule metabolized, also has many hexose transporters (9). This may reflect the varied niches in which C. albicans thrives, which likely offer many different sugars as carbon sources.
Little is known about how C. albicans senses and responds to sugars. A sugar-sensing and signaling pathway that primarily regulates glucose transport has been elucidated in the yeast S. cerevisiae (13, 46). This pathway uses two glucose sensors in the cell membrane—Snf3 and Rgt2—that govern sugar acquisition by regulating the expression of genes encoding hexose transporters (HXTs) (21). The glucose sensors are orthologues of transporters but have long cytoplasmic tails involved in intracellular signaling that are missing from glucose transporters. Sensors cannot transport sugars. Rather, they generate intracellular signals in response to glucose that induce the expression of genes encoding hexose transporters (39). A single missense mutation in the S. cerevisiae glucose sensors illustrates this well: the Snf3(R229K) or Rgt2(R231K) mutations cause constitutive expression of genes encoding hexose transporters, presumably because these mutations convert the sensors into their glucose-bound (signaling) conformations (40).
C. albicans possesses an orthologue of the Snf3 and Rgt2 glucose sensors: Hgt4 (orf19.5962). We present evidence that Hgt4 is a high-affinity glucose sensor that generates an intracellular signal to induce expression of certain HGT genes encoding hexose transporters and that this is required for growth on fermentable carbon sources, for filamentation, and for optimal virulence.
MATERIALS AND METHODS
Yeast strains.
A wild-type C. albicans strain SC5314 (12) was purchased from ATCC (catalogue number MYA-2876). The C. albicans parental strain we used for all gene knockouts is BWP17 (ura3::imm434/ura3::imm434 arg4::hisG/arg4::hisG his1::hisG/his1::hisG) (55); the DAY286 strain is BWP17 that was rendered Arg+Uri+ (44); both strains were gifts from Aaron P. Mitchell (Columbia University). The C. albicans GPR1 parental strain, GPR1/gpr1 heterozygote, gpr1/gpr1-null mutant (gpr1Δ), and gpa2/gpa2-null mutant (gpa2Δ) have been described previously (34), and were obtained from Joseph Heitman (Duke University). The S. cerevisiae snf3Δ rgt2Δ mutant strain (YM6870, similar to YM6370 [22]) was generated by Aneta Kaniak (unpublished data).
Plasmids.
The UAU1 cassette (pBME101) and HIS1-complementing plasmid pDDB78 were gifts from Aaron Mitchell (Columbia University) (10, 51). The HXT1-lacZ reporter (BM3212) was generated by Sabire Ozcan (41). The SAT1 flipper cassette (pSFS2A) was a gift from Joachim Morschhauser (Institute Fur Molekulare Infektions Biologie) (43).
Gene deletions. (i) hgt4Δ.
The UAU1 cassette replaced the HGT4 open reading frame (ORF) as follows. HGT4, including 0.9 kb of DNA upstream and downstream of the ORF, was amplified by the PCR (primers OM5073 and OM5076) and cloned via “gap-repair” (38) into pRS316 (CEN, URA3 (49) in S. cerevisiae. The resulting HGT4 plasmid (BM4913) was linearized with BsrGI, and the PCR-amplified UAU1 cassette (primers OM6040 and OM6042) was inserted by gap repair of pBM4914 into S. cerevisiae, selecting for Ura+ (vector) and Arg+ (full UAU1 insert). The hgt4::UAU1 fragment was released from the plasmid by digesting it with NotI-SalI and used to transform strain BWP17. Arg+ colonies were screened for the Uri+ phenotype. Three independent clones were verified by PCR for the absence of the HGT4 ORF and the presence of UAU1 at hgt4.
(ii) hgt12Δ.
The strategy described above was used with the following modifications. Primers OM5075 and OM5078 amplified the HGT12 ORF plus 0.9 kb of flanking sequence; primers OM6044 and OM6046 amplified the UAU1 cassette. Cloned HGT12 was linearized with HpaI to replace its ORF with UAU1. The hgt12::UAU1 fragment was recovered by digestion of the plasmid with KpnI-SacI and introduced into BWP17 cells.
(iii) Complementing clones.
The HGT4 ORF (primers OM6132 and OM6133) or the HGT4-1 allele (R167K) (primers OM6132 and OM6133, along with OM6134 and OM6135, which encode the R167K mutation) and 1 kb of upstream flanking sequence (“promoter”) and 0.5 kb of downstream flanking sequence (3′-untranslated region [3′UTR]) were amplified by PCR using high-fidelity pHusion polymerase (New England Biolabs). The products (one fragment for HGT4 and two fragments for HGT4-1) were cloned by gap repair into pDDB78 (51) that had been linearized with NotI-SpeI. The clones were verified by determining their DNA sequence, and the resulting plasmids were linearized with NruI for transformation into hgt4Δ strains, selecting for His+ colonies. Controls were the pDDB78 empty vector (“vector”) or pDDB78 containing only the 1 kb upstream and 0.5 kb downstream (relative to the coding ORF) DNA sequences but lacking any coding ORF (“UTRs”). Since the HIS1 locus is on chromosome 5, and BWP17 is heterozygous in this region, all complemented strains were genotyped to determine complementation site. All insertions complemented the wild-type (full-length) chromosome 5a. Two of the three confirmed hgt4Δ mutants were complemented each with five independent wild-type HGT4 clones. Similar results were observed at 30 and 37°C. We investigated whether the HGT4 locus was heterozygous by DNA alignment of orf19.5962 and its allele orf19.13383 (2). Alignment of the 2.2470-kb coding region, including 1 kb upstream and 1 kb downstream of the ORF showed the “alleles” to be identical.
(iv) rgt1Δ.
the SAT1 cassette was used to replace both copies of the RGT1 ORF as follows. Two clones containing either 0.5 kb of DNA from the 5′ and 3′UTRs of RGT1 (clone pBM4707) or 0.5 kb of DNA internal to the RGT1 ORF (0.5 kb from the 5′ end of the gene and 0.5 kb from the 3′ end) (clone pBM4864) were constructed as described below. In each clone, DNA fragments from the RGT1 locus flanked the SAT1 flipper cassette (43) and contained KpnI and SacI restriction sites enabling release of the RGT1 knockout cassettes by KpnI/SacI digest. To create an RGT1/rgt1 heterozygous strain in an hgt4Δ background, strain CM9 was transformed with ∼1 μg of KpnI/SacI-digested pBM4707, and integration of the knockout cassette was selected for by plating transformants on yeast extract-peptone-dextrose (YPD) containing 100 mg of nourseothricin (Nat)/liter. NatR colonies were purified and then grown in YP broth containing 2% maltose to induce loss of the SAT1 cassette through recombination (43). Cells were then plated on YPD, screened for Nat sensitivity, colony purified, and genotyped by PCR to confirm loss of one copy of RGT1. Next, the hgt4Δ, RGT1/rgt1 strain was transformed with ∼1 μg of KpnI/SacI-digested pBM4864 and processed as just described to obtain an hgt4Δ rgt1Δ double-knockout strain. This strain was genotyped by PCR to confirm the presence of both rgt1 deletions and the absence of the wild-type RGT1 gene.
Media.
Standard YPD medium (10 g of yeast extract, 20 g of peptone, 20 g of glucose, and 18 g of Difco agar per liter) was supplemented with 80 to 200 mg of uridine/liter. For SAT1-flipper-mediated gene deletions, 100 mg of nourseothricin/liter was added to the YPD as needed (35). Standard synthetic complete medium (1.7 g of yeast nitrogen base, 5 g of ammonium sulfate, 2 g of −Leu amino acid dropout mix [U.S. Biologicals D9525], 5 ml of 200× leucine, and 18 of g Difco agar/liter) was supplemented with arginine (80 mg/liter), uridine (80 mg/liter), and histidine (40 mg/liter). Spider medium was prepared as previously described (29) and supplemented with arginine, uridine, and histidine (final concentrations as described above). Antimycin A (50 to 75 μl of stock: 1 mg/ml in ethanol) was spread onto petri plates with 30 ml of YPD-uridine (YPD+uri) solid medium.
Yeast culture conditions.
For observing growth on solid medium containing various carbon sources, fresh colonies harvested from YPD+uri plates were resuspended in liquid YPD+uri and grown for 5 h at 30°C, reaching an optical density at 600 nm (OD600) of 2 to 3. Cells were diluted to an OD600 of 0.0001 (ca. 1 CFU/μl) in sterile water, and 20 μl was spotted onto the plates. For liquid cultures (sugar inductions), cells were grown to log phase in synthetic complete medium with 5% glycerol as a carbon source and then diluted back to an OD600 of 0.2 to 0.3 in synthetic complete medium with the indicated carbon sources.
RT-PCR.
Cells grown to log phase in synthetic complete medium were diluted to an OD600 of 0.2 to 0.3 in synthetic complete medium with the indicated carbon sources and grown for 2.5 h at 30°C. Cell pellets were snap-frozen in liquid nitrogen and stored at −80°C. For total RNA purification, cells were resuspended in 400 μl of Tris-EDTA plus 0.5% sodium dodecyl sulfate and 500 μl of hot acid-phenol (65°C) and kept at 65°C for 1 h, with vortexing for 10 s every 10 min. The RNA was phenol-chloroform extracted and ethanol precipitated. A total of 5 μg of RNA was treated with DNase (Ambion DNA-free, catalogue no. 1907), and 500 ng was used in a 20-μl reverse transcription (RT) reaction (Superscript II; Invitrogen). One microliter of cDNA was used in each 20-μl PCR; the number of amplification cycles ranged from 18 (ACT1, HGT7) to 22 (HGT12, HXT10) to 24 (HGT4).
Microarray analysis.
DNA oligonucleotide microarrays were generated that represent 6,346 of the 6,354 predicted ORFs in the annotated C. albicans genome assembly 19 (8). Each ORF is represented by a specific 70-mer oligonucleotide, and one genome equivalent was spotted three times per slide (i.e., n = 3 for each hybridization). Oligonucleotide sequences were selected by using an ArrayOligoSelector (7) and were obtained from Illumina (San Diego, CA). HGT4-1 (CM36) or hgt4Δ (CM32) cells were grown to log phase in synthetic complete medium plus 5% glycerol. Total RNA was prepared as described above. DNA contamination was removed by using RNeasy columns (QIAGEN), and purified RNA was quantified by using a Nanodrop spectrophotometer. The quality was confirmed by using an Agilent 2100 bioanalyzer. A two-step protocol was used for hybridization (3DNA array 350 detection system; Genisphere, Hatfield, PA). First, oligonucleotide arrays were hybridized to the cDNA probes in 2× formamide-based hybridization buffer overnight at 43°C and washed in 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)-0.2% sodium dodecyl sulfate according to the manufacturer's protocol. Fluorescent Cy3- and Cy5-capture reagents were combined in hybridization buffer and added to each array, which were incubated and washed again as described above. Slides were scanned immediately after hybridization on a ScanArray express HT scanner (Perkin-Elmer) to detect Cy3 and Cy5 fluorescence. The laser power was kept constant, and photomultiplier tube (PMT) values were set for optimal intensity with minimal background. An additional scan was done for each slide with the PMT such that <1% of the elements are saturated in order to characterize spots which were saturated at the higher PMT setting. Gridding and analysis of images was performed with ScanArray software express V2.0 (Perkin-Elmer). Significant expression changes were determined as follows: the hybridization intensity values are imported into GeneSpring, and the local background intensity was subtracted. The mean signal and control intensities of the duplicate (or triplicate) spots were calculated. A Lowess curve was fit to the log-intensity versus log-ratio plot. A total of 20.0% of the data was used to calculate the Lowess fit at each point. The resulting curve was used to adjust the control value for each measurement. If the control channel was lower than 10 relative fluorescence units, then 10 was used instead. The mean signals to Lowess-adjusted controlled ratios were calculated. The cross-chip averages were derived from the antilog of the mean of the natural log ratios across the two arrays (original and dye-swap). The data was then filtered as follows. Oligonucleotides that received a “present” call (intensity of >200 relative fluorescence units or local signal-to-background value of >2) by the ScanArray software in two of the four high-PMT scans in either the Cy3 or Cy5 were identified for each condition, and all others were excluded from the analysis. The resulting data set (present genes) was filtered for genes which in either replicate 1 or 2 had (i) a >2-fold change in either the high- or low-PMT scan and (ii) P < 0.05 in either the high- or low-PMT scan. Genes that met these criteria in both replicates are shown.
Mouse procedures.
All animal experimental procedures were approved by the Washington University Institutional Animal Care and Use Committee and by the Animal Studies Committee. Female 5- to 6-week-old BALB/c mice were purchased from Harlan Bioproducts, Indianapolis, IN, and acclimated (five mice/cage) in a pathogen-free facility for 4 weeks. Two independent infection experiments were performed with 10- and 12-week-old mice. HGT4 infections (n = 11 and n = 7, respectively; strain CM87 genotype hgt4::ARG4/hgt4::URA3, HIS1::pDDB78/HGT4 [including 1 kb upstream and 0.5 kb downstream and the coding ORF]) and hgt4Δ infections (n = 9 and n = 8, respectively [strain CM137 genotype hgt4::ARG4/hgt4::URA3, HIS1::pDDB78 carrying only noncoding regions, i.e., 1 kb upstream plus 0.5 kb downstream, of HGT4]) were performed by injecting 7.5 × 105 cells into the lateral tail vein. Cells were prepared by growth to log phase in YPD+uri media and washed thoroughly with endotoxin-free phosphate-buffered saline (PBS), and cell densities were measured by cell counts obtained by using a hemacytometer. Cells were diluted to 3.75 × 106 per ml in PBS, and 200 μl of the suspension was used per infection. Mice determined to be moribund were sacrificed by CO2 asphyxiation, and the death was recorded as the next day. Kidneys were harvested using sterile techniques and Dounce homogenized in 3 ml of sterile PBS, and serial 10-fold dilutions were plated on YPD+uri medium.
RESULTS
A glucose sensor orthologue in C. albicans.
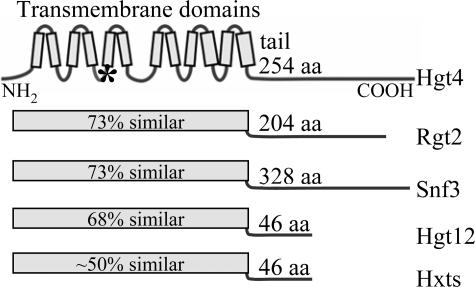
Among the 21 hexose transporter orthologs encoded in the C. albicans genome (11), Hgt4 is most similar to the Snf3 and Rgt2 glucose sensors of S. cerevisiae (Fig. 1). Moreover, because it possesses a long C-terminal cytoplasmic tail, a hallmark of glucose sensors, it is a good candidate for a glucose sensor (Fig. 1). Hgt4 is 56% identical and 73% similar to Snf3 and Rgt2 throughout its predicted transmembrane domains (the glucose-binding region), but the sequences of their cytoplasmic C-terminal tails are not similar. The next most similar protein to Snf3 and Rgt2 is Hgt12 (orf19.7094 or CaSNF32), which has been dubbed a glucose sensor (32), but its short C-terminal tail suggests that it is a sugar transporter rather than a sensor.
FIG. 1.
Glucose sensors and transporters. The C. albicans Hgt4 and the S. cerevisiae Snf3 and Rgt2 glucose sensors are illustrated. For comparison, the C. albicans putative hexose transporter Hgt12 and a composite representation of the S. cerevisiae hexose transporter family (Hxts) are shown. The 12-transmembrane glucose-binding domain and the C-terminal cytoplasmic tails are diagrammed. The percent values indicate similarity to Hgt4 throughout the transmembrane domain. An asterisk indicates the position of the arginine-to-lysine mutation that causes constitutive (sugar-independent) signaling of Snf3(R229K), Rgt2(R231K), or Hgt4(R167K) (40).
Elucidating the function of the C. albicans Hgt4 and Hgt12 proteins.
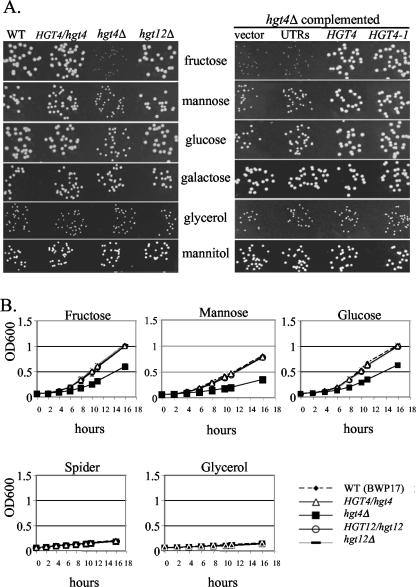
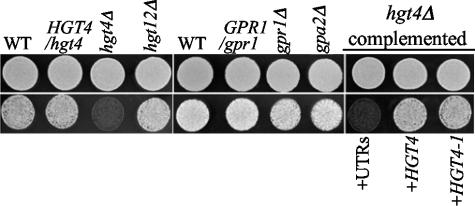
If Hgt4 is indeed a glucose sensor, we would expect it to be required for normal sugar sensing and acquisition; if Hgt12 is indeed simply a glucose transporter, then it is not expected to be required for growth of cells on glucose due to the redundancy of the HGT genes. We constructed homozygous deletion mutants of HGT4 and HGT12 using the UAU1 disruption cassette (see Materials and Methods) (10). The hgt12Δ mutants have no defects in utilization of any of the carbon sources tested, but the hgt4Δ mutants have clear growth defects on solid media with fructose as carbon source, and a slight growth defect on medium containing mannose or glucose that is only apparent at low (0.2%) sugar concentrations (Fig. 2A and data not shown). No hgt4Δ growth defects are observed in highly aerated liquid cultures with 0.2 or 2.0% sugars (data not shown) but, if the cultures are not aerated by shaking, a condition that demands increased fermentation, then the hgt4Δ mutants have a clear growth defect, even at high sugar concentrations (Fig. 2B). Hgt4 is also required for growth on glucose in the presence of antimycin A, which substantially inhibits respiration (Fig. 3) (14, 24). Mutants defective in another glucose sensing pathway triggered by the Gpr1 G-protein coupled receptor and its G protein alpha Gpa2 appear to ferment glucose normally (Fig. 3) (33, 34). These hgt4Δ phenotypes are indeed due to the deletion of the HGT4 ORF because they are complemented by the wild-type HGT4 gene or by a constitutively signaling version of HGT4 (HGT4-1; see below). Thus, Hgt4 is essential for normal growth on low levels of fermentable carbon sources and is especially important for growth when respiration is reduced. Furthermore, these results imply that C. albicans is a facultative anaerobe that can ferment hexoses if necessary, provided that Hgt4 is functional.
FIG. 2.
(A) HGT4 is required for optimal growth on fermentable carbon sources. C. albicans strains were grown 4 to 6 days in 0.2% of various carbon sources at 30°C. In the left panel, WT is the parental strain BWP17 (CM50), HGT4/hgt4 (CM4) is the sensor heterozygote, hgt4Δ is a hgt4/hgt4 knockout strain (CM9), and hgt12Δ is a hgt12/hgt12 knockout strain (CM64). In the right panel, the hgt4Δ strain carrying an empty vector (pDDB78, strain CM32), the “UTRs” only (pDDB78 containing 1 kb of the HGT4 promoter fused to 0.5 kb of the HGT4 3′UTR but lacking any ORF, strain CM137), HGT4 (pDDB78 containing one copy of the wild-type HGT4 gene, strain CM87), and HGT4-1 (pDDB78 containing one copy of the Hgt4 [R167K] allele, strain CM36). (B) HGT4 is essential for growth of C. albicans strains in hypoxic conditions. Log-phase cells prepared in aerated glycerol cultures were inoculated into various 2% carbon sources and grown in static (not shaken) microtiter plates at 30°C. Every 2 h, plates were lightly vortexed, and the optical densities (OD600) of the wells were measured.
FIG. 3.
HGT4 is essential for the growth of C. albicans strains when respiration is inhibited. Cells spotted onto YPD+uri without antimycin A (top row; 3 days growth) or with 2 to 4 μg of antimycin A/ml (bottom row; 5 days of growth). Similar results were observed at 30 and 37°C. For the Hgt4 pathway, WT is the parental strain BWP17 (CM50), HGT4/hgt4 (CM4) is the sensor heterozygote, hgt4Δ is a hgt4/hgt4 knockout strain (CM9), and hgt12Δ is a hgt12/hgt12 knockout strain (CM64). For the Gpr1 pathway, WT is the parental CM149 strain, GPR1/gpr1 is the sensor heterozygote (CM152), gpr1Δ is a gpr1/gpr1 knockout strain (CM151), and gpa2Δ is a gpa2/gpa2 knockout strain (CM150). For hgt4Δ strain complementation, “UTRs” indicates the noncoding flanking regions of HGT4 (CM137), HGT4 indicates one copy of the wild-type HGT4 gene (CM87), HGT4-1 and indicates one copy of the constitutively signaling HGT4 gene (CM35).
Hgt4 contributes to the filamentation of C. albicans cells.
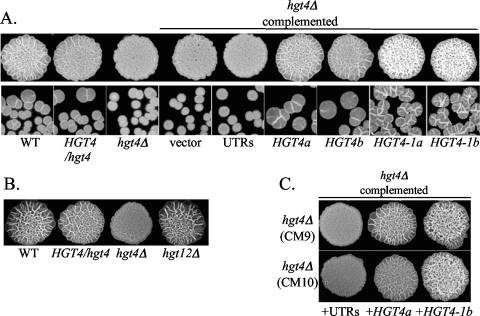
Yeast-form C. albicans cells respond to certain environmental cues such as high pH (>7.0), high temperature (37°C), exposure to serum, engulfment by macrophages, or growth in rich spider medium by forming germ tubes that elongate into branched hyphae or filaments (52). Glucose was recently identified as the main component of serum that is responsible for inducing the filamentation response in C. albicans (15). On solid spider medium (which robustly induces germ tubes and hyphae (filamentation), filamented cells form wrinkled colonies, whereas yeast cells form smooth colonies (29). The hgt4Δ mutant, but not the hgt12Δ mutant, is impaired in its ability to form filaments on spider medium, and filamentation was restored by one copy of the wild-type HGT4 gene (Fig. 4). Interestingly, complementing the hgt4Δ mutant with the constitutively signaling HGT4-1 allele not only rescued the null phenotype but also caused hyper-filamentation of cells (Fig. 4C). Cells harvested from the spider agar plates and stained with calcofluor white (a fluorescent dye that binds to fungal cell walls) showed that the hgt4Δ mutant cells were predominantly yeast form, the HGT4-1-expressing cells were predominantly filamented, and the HGT4-expressing cells produced a mixture of yeast and filamented cells (data not shown). The HGT4-1 mutation changes arginine 167 to lysine; the orthologous change in Rgt2 and Snf3 of S. cerevisiae causes constitutive glucose signaling and constitutive expression of genes regulated by the glucose sensors (Fig. 1) (40). In contrast to their behavior on spider media, all strains developed germ tubes and filamented equally well in YPD plus 10% calf serum (data not shown).
FIG. 4.
(A) Filamentation defects of hgt4Δ mutants. Cells were grown to log phase in YPD+uri, diluted 50-fold (patches) or 5,000-fold (single cells) in sterile water, spotted onto spider medium, and grown for 4 to 5 days at 37°C. WT is the parental strain BWP17 (CM50), HGT4/hgt4 (CM4) is the sensor heterozygote, and hgt4Δ is a hgt4/hgt4 knockout strain (CM9). For hgt4Δ strain complementation, “vector” indicates the pDDB78 empty vector (CM32), “UTRs” indicates the noncoding flanking regions of HGT4 (CM137), HGT4 indicates one copy of the wild-type HGT4 gene (a is CM87 and b is CM97), and HGT4-1 indicates one copy of the constitutively signaling HGT4 allele (a is CM35 and b is CM36). (B) Deleting the Hgt12 hexose transporter does not affect filamentation on spider medium. WT is the parental strain BWP17 (CM50), HGT4/hgt4 (CM4) is the sensor heterozygote, hgt4Δ is a hgt4/hgt4 knockout strain (CM9), and hgt12Δ is a hgt12/hgt12 knockout strain (CM64). (C) The constitutively signaling HGT4-1 allele causes hyperfilamentation. Cells were grown to log phase in YPD+uri, diluted 50-fold in sterile water, spotted onto spider medium, and grown for 4 to 5 days at 37°C. Two independent hgt4Δ mutants (CM9 top row and CM10 bottom row) were complemented: “UTRs” indicates the noncoding flanking regions of HGT4 (pBM4819), HGT4 indicates one copy of the wild-type HGT4 gene (pBM4807), and HGT4-1 indicates one copy of the constitutively signaling HGT4 allele (pBM4782). At 30°C, weak, equivalent filamentation was observed for all strains at day 7 or later (data not shown). The same results were observed when using the DAY286 strain (BWP17 rendered Arg+ Uri+) as the “parent” strain.
Identifying target genes regulated by Hgt4.
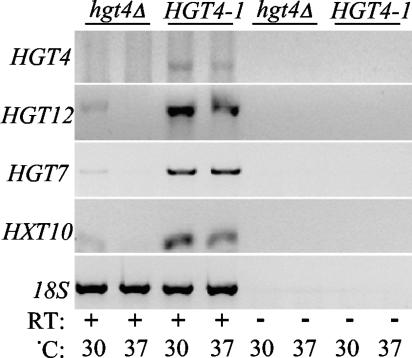
In an attempt to identify genes whose expression is regulated by Hgt4, we profiled the expression of C. albicans genes in HGT4 mutants using an array of DNA oligonucleotides representing nearly all of the ORFs annotated in assembly 19 of the C. albicans genome (8). To avoid detecting transcriptional changes caused by differences between the strains in growth rate, glucose repression, or the yeast-to-hyphal transition, we compared gene expression in the HGT4-1 constitutively signaling mutant to that in an hgt4Δ mutant grown on glycerol. These two mutants are isogenic prototrophs, differing only in their HGT4 allele. Only 35 genes responded differently to the different HGT4 alleles: expression of 28 genes increased, and expression of 7 genes decreased in the HGT4-1 strain compared to the hgt4Δ strain (see Table 2, genes that differed ≥2-fold; also see the supplemental material). Only three genes, all putative hexose transporters, were dramatically induced (>10-fold) by the HGT4-1 mutation: HGT12 (orf19.7094), HGT7 (orf19.2023), and HXT10 (orf19.4384). Cells cultured at 30 and 37° gave nearly identical results. These results suggest that this signaling pathway is primarily dedicated to regulating the expression of genes encoding hexose transporters. HGT4 seems to regulate the expression of only 6 of the 20 HGT genes (see Table 2 and the supplemental material). The differential expression of the three most highly regulated HGT genes was validated by RT-PCR with gene-specific primers (Fig. 5).
TABLE 2.
Genes regulated by Hgt4 (HGT4-1 versus hgt4Δ[ρ])a
| ORF no. | Fold change
|
Gene | GO annotation | |
|---|---|---|---|---|
| 30°C | 37°C | |||
| orf19.5962 | 28.6 | 65.4 | HGT4/SNF3 | High-affinity glucose transport protein; response to glucose stimulus |
| orf19.7094 | 27.9 | 53 | HGT12/SNF32 | Glucose sensor or transporter protein; response to glucose stimulus |
| orf19.2023 | 14.8 | 29.5 | HGT7/HXT3 | Hexose transporter; fructose, mannose, glucose transport |
| orf19.4384 | 13.6 | 29.2 | HXT10 | Fructose symporter; galactose, glucose, mannose transport |
| orf19.508 | 5.83 | 5.5 | QDR1 | Multidrug resistance transporter |
| orf19.5713 | 3.48 | 3.11 | NDE1 | NADH dehydrogenase; cytochrome c oxidase, ethanol fermentation |
| orf19.5730 | 3.14 | 5.28 | orf19.13152 | 3-Polyprenyl-4-hydroxybenzoate decarboxylase |
| orf19.5337 | 3.03 | 3.65 | UBC15 | E2 ubiquitin-conjugating enzyme; ubiquitination, DNA repair |
| orf19.4527 | 2.98 | 2.98 | HGT1 | Hexose transporter |
| orf19.1048 | 2.93 | 2.39 | IFD1 | Benzyl or aryl-alcohol dehydrogenase; aldehyde metabolism |
| orf19.999 | 2.73 | 5.62 | GCA1 | Glucoamylase; alpha glucosidase, cell wall biosynthesis |
| orf19.5911 | 2.71 | 4.8 | CMK1 | Ca2+/calmodulin-dependent protein kinase; signal transduction |
| orf19.3670 | 2.64 | 2.24 | GAL1 | Galactokinase; galactose metabolism, transcription regulation |
| orf19.3392 | 2.61 | 3.04 | DOG1 | 2-Deoxyglucose-6-phosphate phosphatase; glucose metabolism |
| orf19.1509 | 2.47 | 3.37 | ROD1 | Calcium and zinc resistance protein |
| orf19.2244 | 2.46 | 2.78 | orf19.9785 | Aldoketo reductase |
| orf19.2020 | 2.45 | 3.48 | HGT6/HXT6 | Hexose transporter; fructose, mannose, glucose transport |
| orf19.4773 | 2.42 | 3.97 | AOX2 | Alternative oxidase II |
| orf19.4941 | 2.38 | 2.15 | TYE7 | Transcription factor; positive regulator of glycolysis |
| orf19.6803 | 2.36 | 2.74 | HUT1 | UDP-galactose transporter, endoplasmic reticulum translocation, galactosylation |
| orf19.4318 | 2.3 | 2.37 | MIG1 | Transcriptional regulator; transcription corepression, glucose metabolism |
| orf19.542 | 2.26 | 3.32 | HXK2 | Hexokinase II, fructose metabolism |
| orf19.6420 | 2.06 | 2.19 | PGA13 | GPI anchor, similar to mucins, regulated by Tsa1, Cyr1, Nrg1, Tup1, Rlm1 |
| orf19.5816 | 2.02 | 2.14 | EBP7 | NADPH dehydrogenase |
| orf19.2990 | 2 | 2.3 | XOG1 | Glucan 1,3-β-glucosidase; cell wall biogenesis/organization, glucan metabolism |
| orf19.1237 | 1.96 | 1.95 | ARO9 | Aromatic amino acid aminotransferase II; amino acid metabolism |
| orf19.3668 | 1.92 | 4.15 | HGT2 | Hexose transporter |
| orf19.5392 | 1.91 | 4.19 | orf19.12847 | Conserved protein; iron homeostasis siderochrome-iron transport |
| orf19.6993 | 0.44 | 0.47 | GAP2 | General amino acid permease regulated by Nrg1 and Tup1 |
| orf19.5503 | 0.44 | 0.46 | Putative ORF | Hypothetical protein |
| orf19.5975 | 0.42 | 0.5 | orf19.13396 | Zinc-finger transcription factor |
| orf19.4568 | 0.4 | 0.22 | ZCF2 | Zn(2)-Cys(6) transcription factor |
| orf19.2292 | 0.37 | 0.48 | OPT4 | Oligopeptide transporter protein |
| orf19.3902 | 0.31 | 0.42 | orf19.11383 | Hypothetical protein |
| orf19.3904 | 0.3 | 0.4 | orf19.11385 | Hypothetical protein |
The HGT4-1 (CM36) and hgt4Δ[ρ] null (CM32) strains were grown to log phase in synthetic complete medium with glycerol at 30°C (n = 2 each strain) and 37°C (n = 2 each strain). Total RNA was prepared and used to probe 70-mer DNA oligonucleotide microarrays representing all of the ORFs annotated in the C. albicans genome assembly 19. Genes changed by >2-fold at 30 and 37°C are shown.
FIG. 5.
RT-PCR confirmation of the three genes most differentially expressed in the HGT4-1 (CM36) versus hgt4Δ (CM32) strains, as suggested by the microarray results in Table 2. The RNA used for the microarray analyses was reverse transcribed and amplified by PCR with the gene-specific primers indicated in Table 1.
Expression of HGT4 and of Hgt4-regulated genes.
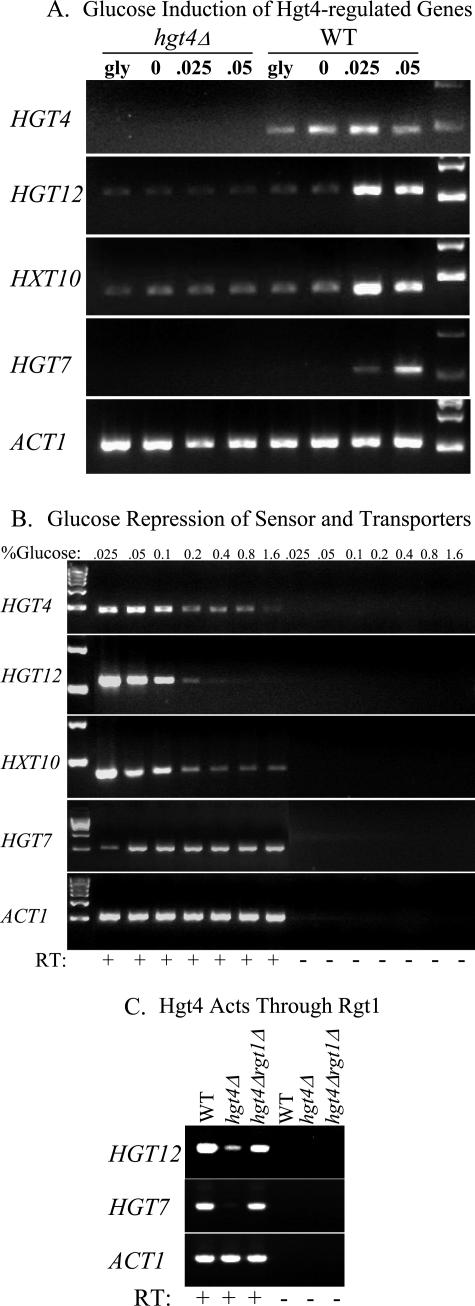
We assessed the expression of HGT4 and its main target genes HGT12, HXT10, and HGT7 in wild-type and hgt4Δ strains (Fig. 6A). HGT4 is expressed in cells grown on a nonfermentable carbon source (glycerol) or on low levels of glucose (Fig. 6A), but its expression is repressed when glucose levels are high (Fig. 6B). The expression of HGT12 and HXT10 transporters is induced by low (physiologic) levels of glucose and repressed by high levels of glucose, suggesting that they encode high-affinity hexose transporters (Fig. 6A and B). HGT12 and HXT10 expression is repressed by high levels of sugar in both wild-type cells and hgt4Δ mutants. In wild-type cells, this occurs via the Mig1 transcriptional repressor, whose expression is induced by the Hgt4 signal (Table 2 and Fig. 6B; also J. A. Sexton et al., unpublished data). In hgt4Δ mutants, HGT12 and HXT10 expression is constitutively repressed via the Rgt1 transcriptional repressor (Fig. 6A and 9; also, data not shown).
FIG. 6.
(A) Glucose induction of HGT12, HXT10, and HGT7 is dependent on HGT4. Cells were grown to log phase (OD600 = 0.5 to 1.0) and then incubated for 2.5 h in 5% glycerol (gly) or with the indicated levels (%) of glucose. Cells were harvested, total RNA was prepared, and RT-PCR was performed on the indicated targets (see Materials and Methods). The strains used were wild-type CM49 and hgt4Δ CM137. Control reactions lacking reverse transcriptase produced no PCR products (data not shown). (B) High levels of glucose repress the expression of HGT4, HGT12, and HXT10 but not HGT7. Cells (CM49) were incubated for 2.5 h with the indicated level of glucose (%). RT-PCR analysis of the indicated target genes is shown. (C) Hgt4 acts through the transcriptional repressor Rgt1. Deleting the RGT1 gene restored the expression of HGT12 and HGT7 in the hgt4Δ mutant. RT-PCR analysis compared the expression of Hgt4-target genes in wild-type DAY286 cells (induced expression) versus hgt4Δ mutant CM9 cells (uninduced expression) versus hgt4Δ rgt1Δ double mutant CM170 cells (constitutive expression). Cells were induced for 2 h in 0.05% glucose. Total RNA was extracted, and RT-PCR was performed as described in the text.
FIG. 9.
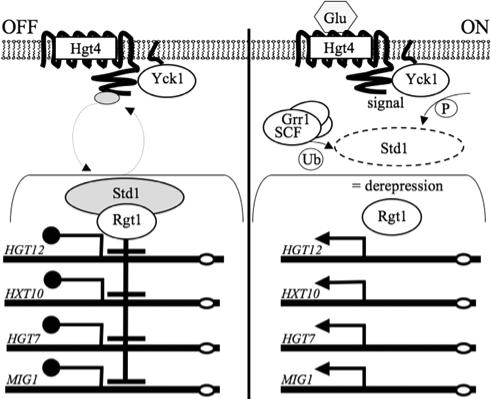
Model for glucose sensing in C. albicans. The Hgt4 glucose sensor resides in the plasma membrane of cells. The C-terminal tail of Hgt4 associates with Std1, a transcriptional corepressor that also associates with the Rgt1 DNA-binding protein. Std1, together with Rgt1, represses expression of the HGT genes. Upon stimulation of the sensor by physiologic levels of glucose, Std1 is phosphorylated by Yck1, then ubiquitinated by SCFGrr1, and finally degraded by the proteasome. Without Std1, Rgt1 is unable to repress transcription, leading to derepression of target genes such as those identified in Table 2.
In contrast, in wild-type cells the expression of HGT7 is induced by glucose but is not repressed by high levels of sugar. Our gene expression profiling experiments also indicate that HGT7 expression is induced in response to high levels of many sugars including glucose, fructose, maltose, and galactose (data not shown). In the hgt4Δ mutants, HGT7 is constitutively repressed (Fig. 6A to 6C and data not shown). Glucose induction of HGT12, HXT10, and HGT7 expression by low levels of glucose (0.025 to 0.1%) requires HGT4, which is consistent with the idea that HGT4 is a high-affinity glucose sensor that controls the expression of downstream genes.
In S. cerevisiae, the intracellular signal generated by the glucose sensors leads to inactivation of the transcriptional repressor Rgt1, resulting in depression of its target genes (see Fig. 9). Deleting the C. albicans RGT1 orthologue (orf19.6173) in the hgt4Δ mutant restored the expression of HGT12, HGT10, and HGT7 to wild-type levels and in fact causes HGT gene expression to be constitutive (Fig. 6C and data not shown). These results are consistent with the view that CaRgt1 is a transcriptional repressor that mediates Hgt4 regulation of the HGT genes.
Determining the glucose responsiveness of Hgt4.
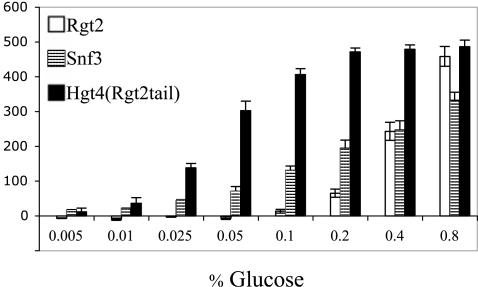
We compared the functionality of Hgt4 to that of the glucose sensors of S. cerevisiae by testing its signaling ability in S. cerevisiae. Because the amino acid sequence of the cytoplasmic (C-terminal) tail of Hgt4 is not similar to that of the tails of the two glucose sensors of S. cerevisiae, we surmised that we needed to provide Hgt4 with a tail that can interact with the components of the glucose-signaling pathway (Mth1 and Std1), so we replaced the C-terminal tail of Hgt4 with the tail of the Rgt2 sensor. The resulting protein (codon optimized for expression in S. cerevisiae) was expressed in a snf3Δ rgt2Δ mutant that carries the HXT1-lacZ gene fusion to report the glucose signaling activity (Table 1). The Hgt4-Rgt2 chimera mediates induction of HXT1 expression in response to low levels of glucose, like the low-glucose (high-affinity) sensor Snf3 (Fig. 7). This is unlikely to simply be due to attaching the Rgt2 tail to Hgt4, because attaching the identical tail to glucose transporters does not bestow upon them the ability to generate a glucose signal (35). This suggests that Hgt4 is a high-affinity glucose receptor. It is notable that the Hgt4-Rgt2tail protein efficiently senses 0.05 to 0.1% glucose, which corresponds to the normal glucose concentration in human blood (0.09% or 5 mM).
TABLE 1.
Reagents used in this study
| Strain, plasmid, or primer | Genotype or sequence | Source or reference |
|---|---|---|
| Strains | ||
| CM49 | SC5314 wild-type (blood isolate) | 12 |
| CM50 | BWP17, ura3::imm434/ura3::imm434 his1::hisG/his1::hisG arg4::hisG/arg4::his | 55 |
| DAY286 | BWP17, ura3::imm434/ura3::imm434 his1::hisG/his1::hisG arg4::his/ARG4::URA3 | 44 |
| CM149 | ura3::imm434/ura3::imm434 TRP1/TRP1::URA3 | 34 |
| CM152 | ura3::imm434/ura3::imm434 GPR1/gpr1::hisG TRP1/TRP1::URA3 | 34 |
| CM151 | ura3::imm434/ura3::imm434 gpr1::hisG/gpr1::hisG TRP1/TRP1::URA3 | 34 |
| CM150 | ura3::imm434/ura3::imm434 gpa2::hisG/gpa2::hisG TRP1/TRP1::URA3 | 34 |
| CM4 | BWP17, HGT4/hgt4::ARG4 | This study |
| CM9 | BWP17, hgt4::ARG4/hgt4::URA3 | This study |
| CM10 | BWP17, hgt4::ARG4/hgt4::URA3 (independently generated null mutant) | This study |
| CM63 | BWP17, HGT12/hgt12::ARG4 | This study |
| CM64 | BWP17, hgt12::ARG4/hgt12::URA3 | This study |
| CM32 | CM9, his1::hisG/HIS1::pDDB78 | This study |
| CM137 | CM9, his1::hisG/HIS1::pDDB78 +HGT4UTRs only (no coding ORF) | This study |
| CM140 | CM9, his1::hisG/HIS1::pDDB78 +HGT4UTRs only (independent transformant) | This study |
| CM87 | CM9, his1::hisG/HIS1::pDDB78 +wtHGT4(a) | This study |
| CM97 | CM9, his1::hisG/HIS1::pDDB78 +wtHGT4(b) independent transformant | This study |
| CM35 | CM9, his1::hisG/HIS1::pDDB78 +HGT4-1(a) | This study |
| CM36 | CM9, his1::hisG/HIS1::pDDB78 +HGT4-1(b) independent transformant | This study |
| CM170 | CM9, rgt1Δ::FRT/rgt1Δ::FRT | This study |
| YM6870 | MATahis3Δ leu2Δ ura3Δ met15Δ LYS2 rgt2::kanMX::natMX snf3::kanMX | 22 |
| YM7309 | YM6870, plasmid pRS316 (CEN, URA3) | This study |
| YM7311 | YM6870, plasmid pBM3272 (pRS316+RGT2) | This study |
| YM7312 | YM6870, plasmid pBM3272 (pRS316+SNF3) | This study |
| YM7314 | YM6870, plasmid pBM4840 (pRS316+HGT4-RGT2tail) | This study |
| Plasmids | ||
| pBM3212 | pYEp367R + HXT1 promoter-lacZ fusion (2μ ori, LEU2 selection) | 40 |
| pBM4665 | pBME101 (UAU1 cassette) | 10 |
| pBM4671 | pDDB78 (HIS1 complementation, TRP1 selection) | 51 |
| pBM4819 | pDDB78 + HGT4 UTRs only (no coding ORF) | This study |
| pBM4807 | pDDB78 + wild-type HGT4(a) | This study |
| pBM4809 | pDDB78 + wild-type HGT4(b) independent clone | This study |
| pBM4780 | pDDB78 + constitutively signaling (R167K) HGT4-1(a) | This study |
| pBM4782 | pDDB78 + constitutively signaling (R167K) HGT4-1(b) independent clone | This study |
| Primersa | ||
| hgt4::UAU1 | ||
| OM5073 | 5′-AACTAGTGGATCCCCCGGGCTGCAGGAATTCtaaattttgttgacagtttttagcattg | This study |
| OM5076 | 5′-CGGTATCGATAAGCTTGATATCGAATTCatgtaatgggtgtataccctttttatc | This study |
| OM6040 | 5′-CTCGTATAATGTACGATGCTTCATTGGAAGACGAGTATTAggttttcccagtcacgacgt | This study |
| OM6042 | 5′-ACCCCTTCTTGTTCCACCACCACTTCCTCTTCCTCTTCATtgtggaattgtgagcggata | This study |
| hgt12::UAU1 | ||
| OM5075 | 5′-AACTAGTGGATCCCCCGGGCTGCAGGAATTCtccagggattaggtgattatttcag | This study |
| OM5078 | 5′-CGGTATCGATAAGCTTGATATCGAATTCttagcggcctgtaacgggactgaatac | This study |
| OM6331 | 5′-TGAGTGCAAATATCCAAGCTCTtagaaggaccacctttgattgtaaatag | This study |
| OM6332 | 5′-ACTAAAAACTATAAACTGTCATTAgggatttggatggtataaacggaaac | This study |
| hgt4Δ complementation | ||
| OM6132 | 5′-gatatcgaattcctgcagcccgggggatccactagtatgatagtgccacttgtcatcatc | This study |
| OM6133 | 5′-atagggcgaattggagctccaccgcggtggcggccgcaactgcccaatgtaatggagttc | This study |
| OM6134b | 5′-gtcgccaaaatggctgaAaggttcagtggtgtttac | This study |
| OM6135b | 5′-gtaaacaccactgaacctTtcagccattttggcgac | This study |
| RT-PCR | ||
| OM6296 | 5′-cattgtataatggcgaagatcag (HGT4 forward) | This study |
| OM6297 | 5′-cagaatcacttggaggatgagc (HGT4 reverse) | This study |
| OM6298 | 5′-tatggtactcaattcttcaagcg (HGT12 forward) | This study |
| OM6299 | 5′-tcatctctaaatgcgtggacac (HGT12 reverse) | This study |
| OM6474 | 5′-gacgttgatgggcagaatcg (HXT10 forward) | This study |
| OM6475 | 5′-ctgtgtatgtcatggccacc (HXT10 reverse) | This study |
| OM6292 | 5′-aaccaagaatctttgaaagggttg (HGT7 forward) | This study |
| OM6295 | 5′-gaaaactaaacatcccataaagacg (HGT7 reverse) | This study |
| OM6306 | 5′-tgatttggctggtagagacttg (ACT1 forward) | This study |
| OM6307 | 5′-tttgtggtgaacaatggatggac (ACT1 reverse) | This study |
Uppercase letters indicate sequences used during gap-repair cloning. Lowercase letters indicate sequences used during PCR amplification.
The uppercase letter indicates the mutation encoding the R167K missense.
FIG. 7.
Signaling by glucose sensors in S. cerevisiae. The snf3Δ rgt2Δ sensor-deficient S. cerevisiae mutant strain (YM6870) carrying a PHXT1-lacZ reporter plasmid (pBM3712) was transformed with constructs expressing wild-type S. cerevisiae Rgt2 (YM7311), wild-type S. cerevisiae Snf3 (YM7312), C. albicans Hgt4 glucose-binding domain fused to the Rgt2 cytoplasmic tail (Hgt4-Rgt2 tail, YM7314), or vector only (YM7309). Values were normalized to cells harboring vector only (baseline). The β-galactosidase activity is indicated in Miller units.
HGT4 contributes to C. albicans virulence.
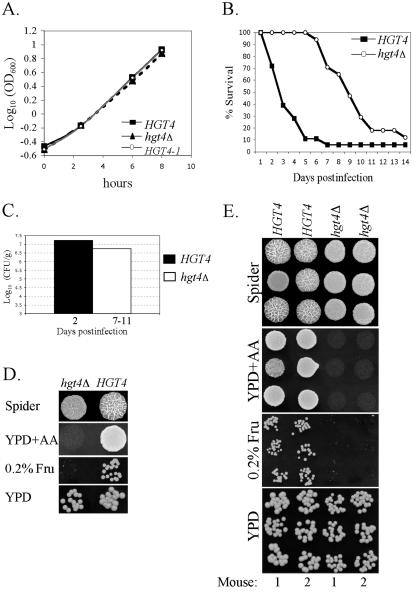
Since Hgt4 is required for filamentation of C. albicans cells and for normal growth on low levels of glucose, we suspected it may be necessary for virulence in vivo. However, since C. albicans favors respiration over fermentation and since growth of the hgt4Δ mutant on glucose is impaired only under conditions in which fermentation is important, it was possible that HGT4 would be irrelevant to virulence. To investigate this, we assessed the colonization and virulence of the hgt4Δ mutant (Fig. 8) (50). We used isogenic, prototrophic strains of C. albicans that differed only in the presence or absence of the HGT4 ORF. Cells used for the infections were prepared from log-phase cultures grown under conditions in which there is no growth rate difference between these strains (Fig. 8A) (36). Cells were injected into the lateral tail veins of female mice, which were monitored for morbidity and mortality. Ninety percent mortality was observed within 5 days for mice infected with the wild-type HGT4 strain but was not observed until 14 days postinfection in mice infected with the hgt4Δ mutant (Fig. 8B). The fungal burden in the kidneys of the mice was determined throughout the course of infection and was similar to that reported in other studies (Fig. 8C) (50). The fungal burdens in hgt4Δ-infected mice were similar to the burdens in mice infected with wild-type cells but, by day 6, 90% of the wild-type-infected mice had died and 90% of the hgt4Δ-infected mice were still alive. This suggests that HGT4 is not necessary for colonization of the kidney. Perhaps the delayed virulence of the hgt4Δ mutant is due to slower organ invasion or to a filamentation defect in vivo.
FIG. 8.
HGT4 is required for optimal virulence in vivo. (A) Growth curves for isogenic C. albicans strains. HGT4 (wild type: CM87 and CM97), hgt4Δ (null mutants: CM137 and CM140), and HGT4-1 (constitutive mutants: CM35 and CM36) cells were grown to log phase in liquid YPD aerated by shaking at 30°C. All standard deviations were <3% of the OD600 values. These C. albicans strains are isogenic and prototrophs, differing only in the HGT4 allele. (B) HGT4 affects virulence during disseminated candidiasis. Mice were injected in the lateral tail vein with 7.5 × 105 cells and monitored for morbidity and mortality. HGT4 indicates the hgt4Δ CM9 complemented with one copy of the wild-type gene, strain CM87 (n = 18), and hgt4Δ indicates the hgt4Δ CM9 complemented with the vector plus noncoding flanking regions of HGT4 (UTRs), strain CM137 (n = 17). (C) The fungal burden was determined for three mice infected with the HGT4 strain (day 2) and for three mice infected with the hgt4Δ strain (one mouse from day 7 and two mice from day 11). The variation between mice was 6 × 106 CFU/g (HGT4 strain) and 6 × 105 CFU/g (hgt4Δ strain). (D) Phenotypes for the original strains used to infect mice in the disseminated Candidiasis experiments in panel C: HGT4 (CM87) and hgt4Δ (CM137). (E) Phenotypes of individual clones recovered postmortem from the kidneys. C. albicans isolates were recovered from two mice infected with the HGT4 strain (day 2) and from two mice infected with the hgt4Δ strain (days 6 and 7). Three independent colonies from each mouse were tested for filamentation on spider medium, for growth on YPD-antimycin A, and for growth on synthetic complete medium with 0.2% fructose.
The sudden onset of mortality seen in the hgt4Δ infection at days 6 to 7 postinfection could have been due to genetic “revertants” in the in vivo yeast population. However, when C. albicans colonies rescued postmortem from the kidneys in two hgt4Δ-infected mice (days 6 to 7 postinfection) and from two HGT4-infected mice (day 2 postinfection) were compared, the hgt4Δ strains had not reverted to the wild-type phenotypes for filamentation on spider medium, growth on YPD plus antimycin A, or growth on 0.2% fructose (Fig. 8D and E). Thus, the hgt4Δ mutants are able to kill mice, albeit more slowly than wild-type cells.
DISCUSSION
C. albicans is a significant contributor to morbidity and mortality for hospitalized and immunocompromised people, in part because of its ability to access and thrive in varied environments. The primary requirement for fitness in any environment is the ability to garner nutrients such as glucose. Previous work has indicated that genes for the HGT4 sensor and the HGT12 transporter increase in expression when C. albicans cells are phagocytosed by macrophages (32, 42) and that these genes are regulated during filamentation, biofilm formation, and in response to antifungal drugs (5, 30), which implies a vital function for these proteins in the host environment.
We have provided evidence that Hgt4 is a sensor of sugar in C. albicans. The Hgt4 sensor appears to sense fructose and mannose, as well as glucose, like the Snf3 and Rgt2 sensors in S. cerevisiae (19, 27, 54). Hgt4 seems to be functionally orthologous to Snf3 of S. cerevisiae, a sensor of low levels of glucose. Like SNF3, HGT4 is expressed when glucose is scarce but not when glucose is abundant, which suggests that both of these proteins are high-affinity glucose receptors. As a high-affinity glucose receptor, C. albicans Hgt4 appears to sense lower glucose concentrations than are sensed by S. cerevisiae Snf3 (Fig. 7). This likely reflects adaptation of these two species to different niches.
The Hgt4 sugar sensor is required for optimal growth on low concentrations of fructose, mannose, and glucose and for utilization of glucose by apparent fermentation. We do not yet understand the reason for the more severe growth defect of the hgt4Δ mutant on fructose compared to mannose and glucose. Hgt4 regulates only 6 of the 20 predicted hexose transporters, and these may be critical for fructose transport, whereas mannose and glucose may be transported by other (constitutively expressed) Hgts. In addition, the first step of hexose metabolism is its phosphorylation catalyzed by hexokinases. C. albicans possesses two hexokinases (Hxk1 and Hxk2) and four glucokinases (Glk1, Glk2, Glk3, and Glk4), only one of which (HXK2) is regulated by Hgt4. Fructose phosphorylation may require Hxk2.
Growth on galactose plates was not affected by the hgt4Δ mutation (Fig. 2A), as expected; in S. cerevisiae, genes encoding enzymes for galactose uptake and utilization are induced by a different signaling pathway (19, 20, 54). A slight growth defect was found for the hgt4Δ mutants in some galactose-based media under certain conditions, but the effect was subtle and inconsistent and may have been due to variable, minute amounts of glucose that often contaminate galactose preparations. It is not surprising that HGT4 is required for fermentation because fermenting cells demand increased glucose transport to achieve the high glucose flux necessary to obtain sufficient energy from that relatively inefficient mode of metabolism. It is not clear to us why the hgt4Δ strains grow more slowly on fructose agar plates when oxygen is abundant and respiration is preferred (Fig. 2A). This could be because (i) colonies on solid media may encounter a microhypoxic environment; (ii) C. albicans, like S. cerevisiae, may ferment sugars even when oxygen is available (37); or (iii) the Hgt4 sensor may regulate genes required for respiration. The third possibility gains some support from our observation that expression of two genes that may influence respiration—the NADH dehydrogenase encoded by CaNDE1/YMX6 (orf19.5713), which transfers electrons from NADH in the cytoplasm to ubiquinone, and orf19.5730, encoding a putative 3-octaprenyl-4-hydroxybenzoate carboxy-lyase involved in ubiquinone biosynthesis—is slightly increased by the HGT4-1 mutation.
HGT4 appears to be important for fermentation because it is required for growth in the presence of antimycin A, which substantially inhibits respiration by blocking electron transfer between cytochromes b and c (14). Wild-type C. albicans is able to carry out some respiration in the presence of antimycin A because it has the alternative oxidases Aox1 (encoded by orf19.4774) and Aox2 (encoded by orf19.4773) that function when cytochromes b and c are inhibited (24) and AOX2 expression is induced by antimycin A (16). AOX2 expression is also induced by the glucose signal generated by Hgt4 (Table 2), and this may account for the severe growth defect of the hgt4Δ mutant on glucose plus antimycin A. The coregulation of fermentative and respiratory pathways via Hgt4 would represent yet another significant transcriptional rewiring that occurred since the divergence of C. albicans and S. cerevisiae (17, 48, 53).
Hgt12 (orf19.7094) is homologous to both Hgt4 and Snf3. It was suggested to be a glucose sensor because it was shown that hgt12 mutations have effects on filamentation of cells (32). We detected no growth or filamentation abnormalities in our hgt12Δ mutants and found that hgt12Δ mutants exhibit normal regulation of the HGT4 target genes HXT10 and HGT7 (data not shown). Thus, we suggest that Hgt12 is likely a hexose transporter rather than a glucose sensor. Another HGT gene (HGT3, orf19.4356) encoding a putative hexose transporter is predicted to possess a C-terminal extension of 177 amino acids, which opens the possibility that another sugar sensor exists in C. albicans.
From these data we propose a model for glucose sensing through Hgt4 in C. albicans that is based on our understanding of the orthologous pathway in S. cerevisiae (Fig. 9). Hgt4 is expressed on the surface of cells growing on nonfermentable carbon sources or at physiologic (relatively low) levels of glucose. Hgt4 likely associates with CaStd1 (orf19.6173), the orthologue of the Std1 and Mth1 transcriptional corepressors of S. cerevisiae. The CaStd1 corepressor probably also associates with CaRgt1 (orf19.2747), a nuclear DNA-binding protein that binds to the promoters of HGT genes encoding glucose transporters and effects their repression. The Hgt4-associated and Rgt1-associated Std1 protein presumably shuttles between the cytoplasm and the nucleus. Upon stimulation of the Hgt4 sensor by glucose, cytoplasmic Std1 becomes phosphorylated by Yck1 (orf19.7001 or orf19.2222), is then ubiquitinated by SCFGrr1(orf19.3944), and is subsequently degraded in the proteasome. Without Std1, the Rgt1 transcriptional repressor is inactive, resulting in derepression of a handful of target genes (identified in Table 2). This model for the C. albicans glucose-sensing and signaling pathway predicts that deletion of RGT1 will suppress the hgt4Δ mutant phenotypes, and that is indeed the case (Fig. 6C and data not shown). The C. albicans and S. cerevisiae glucose-sensing pathways are remarkably similar, with the exception that the Hgt4 glucose signal in C. albicans may affect genes in additional pathways such as drug resistance (QDR1), respiration (NDE1), alternative oxidation (AOX2), and galactose metabolism (GAL1) (Table 2).
At high glucose levels, expression of the genes encoding the high-affinity sensor HGT4 and the transporters HGT12 and HXT10 is repressed. In S. cerevisiae, glucose repression of SNF3 and the transporters HXT2 and HXT4, mediated by the Mig1 and Mig2 repressors, shuts down the high-affinity sensor and transporters when they are not needed. In C. albicans, expression of MIG1 (orthologous to MIG1 and MIG2 in S. cerevisiae) is induced by glucose via the Hgt4 glucose sensor, implying that a feedback repression mechanism exists in C. albicans as well (Table 2). C. albicans genes HGT12 and HXT10 expression mimics S. cerevisiae genes HXT2 and HXT4 (induced by low levels of glucose; repressed by high levels of glucose), and C. albicans gene HGT7 expression behaves like S. cerevisiae gene HXT3 (induced by low and high levels of glucose). Thus, this glucose-sensing pathway functions to ensure that the appropriate glucose transporters are expressed in response to the amount of glucose available.
The most surprising aspect of our results is that HGT4 is essential for optimal virulence in the mouse. This could be simply due to the fact that HGT4 is required for optimal growth in the relatively low concentrations of sugars present in the mammalian bloodstream (<0.2%). We do not favor this idea because aerated cultures of cells exhibit no differences in growth between the hgt4Δ, HGT4, and HGT4-1 strains (growth curves of cells growing on YP, YP plus 5 mM glucose, or synthetic complete media with 0.2% glucose are identical to Fig. 8A [data not shown]). Also, we suspect that C. albicans cells infecting the host spend little time in the bloodstream and thus have little opportunity for growth there.
Oxygen, which is required for respiration, is unlikely to be available to C. albicans cells in the bloodstream because it is sequestered in hemoglobin (18), and it is unclear how available oxygen is to C. albicans cells colonizing the organs of an infected host. We suggest that C. albicans depends at least in part on fermentation during host infection (4, 28); thus, knocking out the Hgt4 sensor reduces virulence.
One of the major reservoirs for C. albicans is the (largely anaerobic) mammalian gastrointestinal tract, a niche where fermentation is undoubtedly important for survival. We have shown that hgt4Δ mutants are severely inhibited for growth in conditions that favor fermentation, even when sugar concentrations are high. Therefore, shutting down fermentation in C. albicans (by inactivating Hgt4) may destroy the ability of this opportunistic pathogen to colonize the human gut. This along with the cell surface location of the glucose sensor makes it an attractive potential drug target.
Supplementary Material
Acknowledgments
We thank Aaron Mitchell and Joseph Heitman for strains, Judith Berman for insightful discussions, and Paul Cliften and Seth Crosby for DNA microarray design and data analysis.
This study was supported by the Pfizer/Washington University Biomedical Research Program Grant and a W. M. Keck postdoctoral fellowship (to J.A.S.).
Footnotes
Supplemental material for this article may be found at http://ec.asm.org/.
REFERENCES
- 1.Andrutis, K. A., P. J. Riggle, C. A. Kumamoto, and S. Tzipori. 2000. Intestinal lesions associated with disseminated candidiasis in an experimental animal model. J. Clin. Microbiol. 38:2317-2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arnaud, M. B., M. C. Costanzo, M. S. Skrzypek, G. Binkley, C. Lane, S. R. Miyasato, and G. Sherlock. 2005. The Candida Genome Database (CGD), a community resource for Candida albicans gene and protein information. Nucleic Acids Res. 33:D358-D363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Backhed, F., R. E. Ley, J. L. Sonnenburg, D. A. Peterson, and J. I. Gordon. 2005. Host-bacterial mutualism in the human intestine. Science 307:1915-1920. [DOI] [PubMed] [Google Scholar]
- 4.Barelle, C. J., C. L. Priest, D. M. Maccallum, N. A. Gow, F. C. Odds, and A. J. Brown. 2006. Niche-specific regulation of central metabolic pathways in a fungal pathogen. Cell Microbiol. 8:961-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barker, K. S., S. Crisp, N. Wiederhold, R. E. Lewis, B. Bareither, J. Eckstein, R. Barbuch, M. Bard, and P. D. Rogers. 2004. Genome-wide expression profiling reveals genes associated with amphotericin B and fluconazole resistance in experimentally induced antifungal resistant isolates of Candida albicans. J. Antimicrob. Chemother. 54:376-385. [DOI] [PubMed] [Google Scholar]
- 6.Bendel, C. M., D. J. Hess, R. M. Garni, M. Henry-Stanley, and C. L. Wells. 2003. Comparative virulence of Candida albicans yeast and filamentous forms in orally and intravenously inoculated mice. Crit. Care Med. 31:501-507. [DOI] [PubMed] [Google Scholar]
- 7.Bozdech, Z., J. Zhu, M. P. Joachimiak, F. E. Cohen, B. Pulliam, and J. L. DeRisi. 2003. Expression profiling of the schizont and trophozoite stages of Plasmodium falciparum with a long-oligonucleotide microarray. Genome Biol. 4:R9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Braun, B. R., M. van Het Hoogqq, C. d'Enfert, M. Martchenko, J. Dungan, A. Kuo, D. O. Inglis, M. A. Uhl, H. Hogues, M. Berriman, M. Lorenz, A. Levitin, U. Oberholzer, C. Bachewich, D. Harcus, A. Marcil, D. Dignard, T. Iouk, R. Zito, L. Frangeul, F. Tekaia, K. Rutherford, E. Wang, C. A. Munro, S. Bates, N. A. Gow, L. L. Hoyer, G. Kohler, J. Morschhauser, G. Newport, S. Znaidi, M. Raymond, B. Turcotte, G. Sherlock, M. Costanzo, J. Ihmels, J. Berman, D. Sanglard, N. Agabian, A. P. Mitchell, A. D. Johnson, M. Whiteway, and A. Nantel. 2005. A human-curated annotation of the Candida albicans genome. PLoS Genet. 1:36-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dumitru, R., J. M. Hornby, and K. W. Nickerson. 2004. Defined anaerobic growth medium for studying Candida albicans basic biology and resistance to eight antifungal drugs. Antimicrob. Agents Chemother. 48:2350-2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enloe, B., A. Diamond, and A. P. Mitchell. 2000. A single-transformation gene function test in diploid Candida albicans. J. Bacteriol. 182:5730-5736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fan, J., V. Chaturvedi, and S. H. Shen. 2002. Identification and phylogenetic analysis of a glucose transporter gene family from the human pathogenic yeast Candida albicans. J. Mol. Evol. 55:336-346. [DOI] [PubMed] [Google Scholar]
- 12.Fonzi, W. A., and M. Y. Irwin. 1993. Isogenic strain construction and gene mapping in Candida albicans. Genetics 134:717-728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gagiano, M., F. F. Bauer, and I. S. Pretorius. 2002. The sensing of nutritional status and the relationship to filamentous growth in Saccharomyces cerevisiae. FEMS Yeast Res. 2:433-470. [DOI] [PubMed] [Google Scholar]
- 14.Helmerhorst, E. J., M. P. Murphy, R. F. Troxler, and F. G. Oppenheim. 2002. Characterization of the mitochondrial respiratory pathways in Candida albicans. Biochim. Biophys. Acta 1556:73-80. [DOI] [PubMed] [Google Scholar]
- 15.Hudson, D. A., Q. L. Sciascia, R. J. Sanders, G. E. Norris, P. J. Edwards, P. A. Sullivan, and P. C. Farley. 2004. Identification of the dialysable serum inducer of germ-tube formation in Candida albicans. Microbiology 150:3041-3049. [DOI] [PubMed] [Google Scholar]
- 16.Huh, W. K., and S. O. Kang. 2001. Characterization of the gene family encoding alternative oxidase from Candida albicans. Biochem. J. 356:595-604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ihmels, J., S. Bergmann, M. Gerami-Nejad, I. Yanai, M. McClellan, J. Berman, and N. Barkai. 2005. Rewiring of the yeast transcriptional network through the evolution of motif usage. Science 309:938-940. [DOI] [PubMed] [Google Scholar]
- 18.Jensen, F. B. 2004. Red blood cell pH, the Bohr effect, and other oxygenation-linked phenomena in blood O2 and CO2 transport. Acta Physiol. Scand. 182:215-227. [DOI] [PubMed] [Google Scholar]
- 19.Jiang, H., I. Medintz, B. Zhang, and C. A. Michels. 2000. Metabolic signals trigger glucose-induced inactivation of maltose permease in Saccharomyces. J. Bacteriol. 182:647-654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnston, M., J. S. Flick, and T. Pexton. 1994. Multiple mechanisms provide rapid and stringent glucose repression of GAL gene expression in Saccharomyces cerevisiae. Mol. Cell. Biol. 14:3834-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Johnston, M., and J. H. Kim. 2005. Glucose as a hormone: receptor-mediated glucose sensing in the yeast Saccharomyces cerevisiae. Biochem. Soc. Trans. 33:247-252. [DOI] [PubMed] [Google Scholar]
- 22.Kaniak, A., Z. Xue, D. Macool, J. H. Kim, and M. Johnston. 2004. Regulatory network connecting two glucose signal transduction pathways in Saccharomyces cerevisiae. Eukaryot. Cell 3:221-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Khatib, R., J. Ramanathan, K. M. Riederer, D. DePoister, Jr., and J. Baran, Jr. 2002. Limited genetic diversity of Candida albicans in fecal flora of healthy volunteers and inpatients: a proposed basis for strain homogeneity in clinical isolates. Mycoses 45:393-398. [DOI] [PubMed] [Google Scholar]
- 24.Kot, E. J., V. L. Olson, L. J. Rolewic, and D. O. McClary. 1976. An alternate respiratory pathway in Candida albicans. Antonie Leeuwenhoek 42:33-48. [DOI] [PubMed] [Google Scholar]
- 25.Kullberg, B. J., and A. M. Oude Lashof. 2002. Epidemiology of opportunistic invasive mycoses. Eur. J. Med. Res. 7:183-191. [PubMed] [Google Scholar]
- 26.Kumamoto, C. A., and M. D. Vinces. 2005. Alternative Candida albicans lifestyles: growth on surfaces. Annu. Rev. Microbiol. 59:113-133. [DOI] [PubMed] [Google Scholar]
- 27.Lagunas, R. 1993. Sugar transport in Saccharomyces cerevisiae. FEMS Microbiol. Rev. 10:229-242. [DOI] [PubMed] [Google Scholar]
- 28.Lan, C. Y., G. Newport, L. A. Murillo, T. Jones, S. Scherer, R. W. Davis, and N. Agabian. 2002. Metabolic specialization associated with phenotypic switching in Candida albicans. Proc. Natl. Acad. Sci. USA 99:14907-14912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu, H., J. Kohler, and G. R. Fink. 1994. Suppression of hyphal formation in Candida albicans by mutation of a STE12 homolog. Science 266:1723-1726. [DOI] [PubMed] [Google Scholar]
- 30.Liu, T. T., R. E. Lee, K. S. Barker, L. Wei, R. Homayouni, and P. D. Rogers. 2005. Genome-wide expression profiling of the response to azole, polyene, echinocandin, and pyrimidine antifungal agents in Candida albicans. Antimicrob. Agents Chemother. 49:2226-2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Long, S. S., and D. K. Stevenson. 2005. Reducing Candida infections during neonatal intensive care: management choices, infection control, and fluconazole prophylaxis. J. Pediatr. 147:135-141. [DOI] [PubMed] [Google Scholar]
- 32.Luongo, M., A. Porta, and B. Maresca. 2005. Homology, disruption, and phenotypic analysis of CaGS Candida albicans gene induced during macrophage infection. FEMS Immunol. Med. Microbiol. 45:471-478. [DOI] [PubMed] [Google Scholar]
- 33.Maidan, M. M., L. De Rop, J. Serneels, S. Exler, S. Rupp, H. Tournu, J. M. Thevelein, and P. Van Dijck. 2005. The G protein-coupled receptor Gpr1 and the Gα protein Gpa2 act through the cAMP-protein kinase A pathway to induce morphogenesis in Candida albicans. Mol. Biol. Cell 16:1971-1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miwa, T., Y. Takagi, M. Shinozaki, C. W. Yun, W. A. Schell, J. R. Perfect, H. Kumagai, and H. Tamaki. 2004. Gpr1, a putative G-protein-coupled receptor, regulates morphogenesis and hypha formation in the pathogenic fungus Candida albicans. Eukaryot. Cell 3:919-931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moriya, H., and M. Johnston. 2004. Glucose sensing and signaling in Saccharomyces cerevisiae through the Rgt2 glucose sensor and casein kinase I. Proc. Natl. Acad. Sci. USA 101:1572-1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Odds, F. C., L. Van Nuffel, and N. A. Gow. 2000. Survival in experimental Candida albicans infections depends on inoculum growth conditions as well as animal host. Microbiology 146(Pt. 8):1881-1889. [DOI] [PubMed] [Google Scholar]
- 37.Ogasawara, A., K. Odahara, M. Toume, T. Watanabe, T. Mikami, and T. Matsumoto. 2006. Change in the respiration system of Candida albicans in the lag and log growth phase. Biol. Pharm. Bull. 29:448-450. [DOI] [PubMed] [Google Scholar]
- 38.Orr-Weaver, T. L., and J. W. Szostak. 1983. Yeast recombination: the association between double-strand gap repair and crossing-over. Proc. Natl. Acad. Sci. USA 80:4417-4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ozcan, S., J. Dover, and M. Johnston. 1998. Glucose sensing and signaling by two glucose receptors in the yeast Saccharomyces cerevisiae. EMBO J. 17:2566-2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ozcan, S., J. Dover, A. G. Rosenwald, S. Wolfl, and M. Johnston. 1996. Two glucose transporters in Saccharomyces cerevisiae are glucose sensors that generate a signal for induction of gene expression. Proc. Natl. Acad. Sci. USA 93:12428-12432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ozcan, S., and M. Johnston. 1995. Three different regulatory mechanisms enable yeast hexose transporter (HXT) genes to be induced by different levels of glucose. Mol. Cell. Biol. 15:1564-1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prigneau, O., A. Porta, J. A. Poudrier, S. Colonna-Romano, T. Noel, and B. Maresca. 2003. Genes involved in beta-oxidation, energy metabolism and glyoxylate cycle are induced by Candida albicans during macrophage infection. Yeast 20:723-730. [DOI] [PubMed] [Google Scholar]
- 43.Reuss, O., A. Vik, R. Kolter, and J. Morschhauser. 2004. The SAT1 flipper, an optimized tool for gene disruption in Candida albicans. Gene 341:119-127. [DOI] [PubMed] [Google Scholar]
- 44.Richard, M. L., C. J. Nobile, V. M. Bruno, and A. P. Mitchell. 2005. Candida albicans biofilm-defective mutants. Eukaryot. Cell 4:1493-1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Richardson, M. D. 2005. Changing patterns and trends in systemic fungal infections. J. Antimicrob. Chemother. 56(Suppl. 1):i5-i11. [DOI] [PubMed] [Google Scholar]
- 46.Santangelo, G. M. 2006. Glucose signaling in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 70:253-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Saville, S. P., A. L. Lazzell, C. Monteagudo, and J. L. Lopez-Ribot. 2003. Engineered control of cell morphology in vivo reveals distinct roles for yeast and filamentous forms of Candida albicans during infection. Eukaryot. Cell 2:1053-1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scannell, D. R., and K. Wolfe. 2004. Rewiring the transcriptional regulatory circuits of cells. Genome Biol. 5:206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sikorski, R. S., and P. Hieter. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122:19-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Spellberg, B., A. S. Ibrahim, J. E. Edwards, Jr., and S. G. Filler. 2005. Mice with disseminated candidiasis die of progressive sepsis. J. Infect. Dis. 192:336-343. [DOI] [PubMed] [Google Scholar]
- 51.Spreghini, E., D. A. Davis, R. Subaran, M. Kim, and A. P. Mitchell. 2003. Roles of Candida albicans Dfg5p and Dcw1p cell surface proteins in growth and hypha formation. Eukaryot. Cell 2:746-755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sudbery, P., N. Gow, and J. Berman. 2004. The distinct morphogenic states of Candida albicans. Trends Microbiol. 12:317-324. [DOI] [PubMed] [Google Scholar]
- 53.Tsong, A. E., M. G. Miller, R. M. Raisner, and A. D. Johnson. 2003. Evolution of a combinatorial transcriptional circuit: a case study in yeasts. Cell 115:389-399. [DOI] [PubMed] [Google Scholar]
- 54.Wieczorke, R., S. Krampe, T. Weierstall, K. Freidel, C. P. Hollenberg, and E. Boles. 1999. Concurrent knockout of at least 20 transporter genes is required to block uptake of hexoses in Saccharomyces cerevisiae. FEBS Lett. 464:123-128. [DOI] [PubMed] [Google Scholar]
- 55.Wilson, R. B., D. Davis, and A. P. Mitchell. 1999. Rapid hypothesis testing with Candida albicans through gene disruption with short homology regions. J. Bacteriol. 181:1868-1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wisplinghoff, H., T. Bischoff, S. M. Tallent, H. Seifert, R. P. Wenzel, and M. B. Edmond. 2004. Nosocomial bloodstream infections in US hospitals: analysis of 24,179 cases from a prospective nationwide surveillance study. Clin. Infect. Dis. 39:309-317. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.