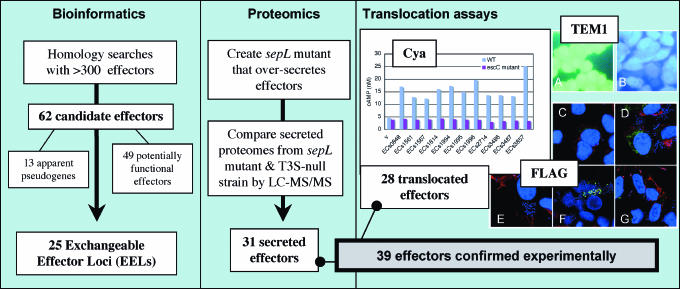
Fig. 1.
Experimental flow chart. BLAST searches with a comprehensive database of known and predicted effector sequences were used to identify 62 candidate effectors in 25 loci within the genome of EHEC O157:H7 Sakai. Using a proteomic approach, 31 candidate effectors were found to be secreted by T3S. Subsequently, three methods for measuring T3S-dependent translocation into eukaryote cells were used to identify 28 translocated effectors. In total, 39 candidate effectors were confirmed experimentally either by proteomics or by translocation assays or both methods. Representative translocation data (including controls) are provided for Cya, TEM1 and FLAG translocation assays. Cya: cAMP levels of Caco-2 cell extracts after infection with wildtype (WT) or escC− (T3S negative) E. coli carrying CyaA-effector fusion plasmids (v = vector only); TEM1: HeLa cell fluorescence was observed after infection with WT or escN− (T3S negative) E. coli carrying TEM1-effector fusion plasmids. Blue or green fluorescence indicates translocation or no translocation, respectively. (A) ECs1567 (escN− mutant). (B) ECs1567 (WT); FLAG: Caco-2 cell fluorescence was observed after infection with WT E. coli carrying FLAG-effector fusion plasmids. FLAG-effector fusion proteins, nucleus and F-actin were fluorescently stained green, blue, and red, respectively. Green fluorescence within Caco-2 cells indicates translocation. (C) ECs1567. (D) ECs1814. (E) Vector only. (F) ECs1994.(G) ECs3485. Further details of the experimental approach are described in Results and Discussion.