Abstract
d-Serine is an amino acid present in mammalian urine that is inhibitory to Escherichia coli strains lacking a functional dsdA gene. Counterintuitively, a dsdA strain of E. coli clinical isolate CFT073 hypercolonizes the bladder and kidneys of mice relative to wild type during a coinfection in the murine model of urinary tract infection. We are interested in the mechanisms for uptake of d-serine in CFT073. d-Serine enters E. coli K-12 via CycA, the d-alanine transporter and d-cycloserine sensitivity locus. CFT073 cycA can grow on minimal medium with d-serine as a sole carbon source. The dsdX gene of the dsdCXA locus is a likely candidate for an additional d-serine transporter based on its predicted amino acid sequence similarity to gluconate transporters. In minimal medium, CFT073 dsdX can grow on d-serine as a sole carbon source; however, CFT073 dsdX cycA cannot. Additionally, CFT073 dsdXA cycA is not sensitive to inhibitory concentrations of d-serine during growth on glycerol and d-serine minimal medium. d-[14C]serine uptake experiments with CFT073 dsdX cycA harboring dsdX or cycA recombinant plasmids confirm that d-serine is able to enter E. coli cells via CycA or DsdX. In whole-cell d-[14C]serine uptake experiments, DsdX has an apparent Km of 58.75 μM and a Vmax of 75.96 nmol/min/mg, and CycA has an apparent Km of 82.40 μM and a Vmax of 58.90 nmol/min/mg. Only d-threonine marginally inhibits DsdX-mediated d-serine transport, whereas d-alanine, glycine, and d-cycloserine inhibit CycA-mediated d-serine transport. DsdX or CycA is sufficient to transport physiological quantities of d-serine, but DsdX is a d-serine-specific permease.
Escherichia coli is a normal resident of the vertebrate large intestine, and certain pathogenic strains are capable of infecting sites outside of the intestine. The sequencing of multiple E. coli genomes has allowed for a better insight into what genes may permit success in niches outside the intestine. Comparison of an E. coli K-12 isolate (MG1655) to an O157:H7 enterohemorrhagic E. coli isolate (EDL933) and a uropathogenic E. coli (UPEC) isolate (CFT073) showed numerous genetic differences between the strains. Less than 50% of the apparent genes were common to all three strains; these common genes represent what can be thought of as the core chromosome of E. coli. The variable regions specific to each isolate include genes potentially important to disease or growth in different environments (38). One such variable site is a genetic island starting 3′ to the argW tRNA gene and ending with the dsdCXA locus. The dsdCXA locus is intact in MG1655 and CFT073 but is truncated in EDL933 (28). In fact, the dsdCXA locus is intact in most extraintestinal pathogenic E. coli (ExPEC) strains but is truncated in the vast majority of diarrheagenic E. coli strains (R. L. Moritz and R. A. Welch, submitted for publication). This region of the E. coli chromosome frequently replaces dsdC and dsdX with csrRAKB genes responsible for sucrose catabolism (12, 28).
The E. coli dsdCXA locus permits growth on d-serine as a sole carbon and nitrogen source. The E. coli K-12 dsdCXA locus was extensively studied by McFall and coworkers (23). The DNA sequence of the locus was originally described by this group, but apparent sequence assembly problems and the inability at the time to create targeted, site-specific mutations prevented proper identification and functional analysis of dsdX. The dsdC gene encodes a Lys-R-type transcriptional regulator that induces transcription of dsdX and dsdA in the presence of d-serine and inhibits its own transcription in the absence of d-serine (23). DsdX has been hypothesized to act as a d-serine transporter (23). The dsdA gene encodes a pyridoxal phosphate-dependent d-serine deaminase (DsdA) that degrades d-serine to ammonia and pyruvate (18). d-Serine catabolism is biologically important because d-serine is available in some environments as a readily utilizable nutrient source, but it can also have also inhibitory effects on growth. d-Serine is bacteriostatic to cells lacking DsdA grown in minimal medium (16). d-Serine toxicity on minimal medium can be reversed with expression of functional DsdA or by the addition of pantothenate or β-alanine to the medium; this suggests that the inhibitory effect of d-serine is associated with pantothenate biosynthesis due to the structural similarity between d-serine and β-alanine (2, 4, 7, 15). A d-serine deaminase gene can serve as a selective marker on par with antibiotic resistance genes for bacteria (16), yeast (35), or plant transformations (8) due to the toxicity of d-serine. Despite the fact that d-serine is toxic to many living organisms, d-serine is one of the most prevalent amino acids excreted in mammalian urine, at reported levels of 3 to 40 μg/ml, and it can be found in mammalian blood as well (11, 24). Strains of E. coli residing within the bladder show increased dsdA expression as a result of d-serine present in human urine (29). Additionally, d-serine is found in mammalian brains, where it acts as a glycine coagonist with N-methyl-d-aspartic acid receptors (14, 20, 21, 34). The observation that a functional dsdCXA locus is present in ExPEC strains and the likelihood of ExPEC strains encountering d-serine in the blood, brain, or urinary tract led to the hypothesis that the dsdCXA genes are involved in either ExPEC carbon and nitrogen acquisition or in response to d-serine as an environmental signal in pathogenesis. Compared to the wild type, a CFT073 dsdA mutant is impaired for growth in human urine, is more motile, and exhibits increased colonization levels in the murine bladder and kidneys (28). An increased understanding of the functions of the gene products of the dsdCXA locus will allow for a better interpretation of phenotypes of the CFT073 dsdA mutant.
DsdX has no significant sequence similarity to CycA, the only described d-serine transporter in E. coli. CycA can transport β-alanine, d-alanine, glycine, and d-cycloserine, in addition to d-serine (3, 13, 30, 33, 36, 37). The dsdX and cycA genes display peak expression under different growth conditions. Expression of cycA is controlled by the nitrogen-scavenging system (Nac) under low nitrogen (10, 27, 40), whereas dsdX expression is controlled by DsdC and requires d-serine for chromosomal induction (23). DsdX has an amino acid sequence suggesting it is a membrane transport protein. Specifically, DsdX has 33% identity with GntU, a gluconate transporter in E. coli. Due to its amino acid sequence, DsdX was classified as a member of the gluconate:H+ symporter family (TC 2.A.8) (31) but has been experimentally shown not to transport gluconate (24). Experimental and bioinformatic evidence strongly suggest it is an inner membrane protein (5). We hypothesize that DsdX is a d-serine-specific permease, in part because dsdX is cotranscribed with dsdA in the presence of d-serine (23). Elucidation of the function of DsdX will allow for a more complete understanding of d-serine metabolism in E. coli. We initially investigated d-serine transport mediated by DsdX by examining genetic knockouts of dsdX, dsdA, cycA, and combinations for an ability to grow on different carbon sources. The genetic test supported the hypothesis that DsdX is a second d-serine transporter. Transport of d-serine was confirmed by monitoring uptake of d-[14C]serine by relevant strains from the genetic experiment. Unlabeled amino acids likely to be transported by DsdX were tested for their ability to impair uptake of d-[14C]serine to examine their possible role as additional substrates. Km and Vmax values for DsdX and CycA d-serine transport were also determined, and DsdX was found to be a more efficient transporter of d-serine than CycA.
MATERIALS AND METHODS
Strains and media.
Deletion mutants were created in WAM 2267, E. coli strain CFT073 (ATCC 700928). WAM 2603, E. coli AAEC185, was used as a host for construction of recombinant plasmids.
LB broth and agar were purchased from Fisher Scientific and prepared as per the manufacturer's instructions. Morpholinepropanesulfonic acid (MOPS) minimal medium was prepared as described elsewhere (22), with the substitution of 43.4 mM glycerol for glucose and the omission of thiamine, giving a final composition of 1.32 mM K2HPO4, 9.52 mM NH4Cl, 0.523 mM MgCl2, 0.276 mM K2SO4, 10 μM FeSO4, 0.5 μM CaCl2, 50 mM NaCl, 40 mM MOPS, 4 mM Tricine, 3 nM (NH4)6(MO7)24, 0.4 μM H2BO3, 30 nM CoCl2, 10 nM CuSO4, 80 nM MnCl2, 10 nM ZnSO4, and 43.4 mM glycerol. Additions or substitutions to MOPS minimal medium relative to this base recipe are noted below. MOPS-Tris buffer contained 0.1 M MOPS, 8 mM MgSO4, and 8 mM Tris base, and the pH was adjusted to 7.0 (9). MOPS-Tris survival buffer used for resuspension of cell pellets additionally contained 43.4 mM glycerol for a carbon source. d-[14C]serine was purchased from American Radiolabeled Chemicals; all other amino acids were purchased from Sigma.
Strain construction.
A list of strains is found in Table 1. Mutants with deletions in dsdX, dsdA, and cycA were created using the λ Red recombination method of Datsenko and Wanner (6). Strain WAM 2850 harboring plasmid pKD46 was grown in the presence of 250 μg/ml carbenicillin and 10 mM l-arabinose to select for the plasmid and induce production of the λ Red recombinase, respectively. A PCR-amplified DNA fragment with regions of homology to the gene of interest was electroporated into WAM 2850. PCR fragments were generated using pKD4, creating a kanamycin-resistant fragment, for the dsd deletions; pKD3 was the template for the cycA deletion and had a chloramphenicol resistance marker. Oligos for deletion of dsdX were P0 1118 (5′ TTCAATATCATCAGGTTAATCACAGGGGAAGGTGAGATTGTGTAGGCTGGAGCTGCTTC 3′) and P2 651 (5′ GAATGTGCCCGCCAGAGCGATGACTGAAGCGATAAATGCATATGAATATCCTCCTTAG 3′). For deletion of dsdA, oligos P0 1052 (5′ CCTGCTGTCATTTATCATCTAAGCGCAAAGAGACGTACTTGTGTAGGCTGGAGCTGCTTCG 3′) and P2 1053 (5′ CACCCAGGGAAAGGATGGCGATGCTGCGTTGAAACGTTACATATGAATATCCTCCTTAG 3′) were used. The cycA deletion was generated with P0 693 (5′ CCTGAACAACACAGACAGGTACAGGAAGAAAAAAAACTGTGTAGGCTGGAGCTGCTTC 3′) and P2 694 (5′ AAAGCTGGATGGCATTGCGCCATCCAGCATGATAATGCGACATATGAATATCCTCCTTA 5′). Generation of the CFT073 dsdXA mutant was accomplished with a PCR fragment generated from P0 1118 and P2 1053. Fragment insertion was confirmed with PCR. Once the desired fragment(s) was inserted, plasmid pCP20 was electroporated into the strain to facilitate excision of the FLP sites in the fragment(s). The strain was cured of pCP20 by heat shock at 42°C, and loss of the PCR fragment(s) was confirmed by patching colonies onto selective medium to look for loss of antibiotic resistance. Gene deletions and loss of antibiotic resistance genes were confirmed by PCR.
TABLE 1.
Bacterial strains used in this study
| WAM no. | Strain description | Source or reference |
|---|---|---|
| 2267 | CFT073 wild type | 19 |
| 2603 | E. coli AAEC185 | 1 |
| 2614 | CFT073445 N-terminal base pair dsdA | 28 |
| 2684 | pKD3 host | 6 |
| 2685 | pKD4 host | 6 |
| 2687 | pKD46 host | 6 |
| 2688 | Dh5αa + pCP20 | 6 |
| 2732 | CFT073 cycA | Shai Pellet |
| 2811 | WAM 2614 + pKD46 | Rebecca Moritz |
| 2850 | CFT073 wild type + pKD46 | Peter Redford |
| 2967 | CFT073 dsdXA | This study |
| 2981 | CFT073 dsdX | This study |
| 3147 | CFT073 dsdX cycA | This study |
| 3160 | CFT073 dsdXA cycA | This study |
| 3386 | WAM 2603 + pAAdsdX | This study |
| 3387 | WAM 3147 + pAAdsdX | This study |
| 3388 | WAM 3160 + pAAdsdX | This study |
| 3480 | WAM 2603 + pAAcycA | This study |
| 3481 | WAM 3147 + pAAcycA | This study |
| 3482 | WAM 3160 + pAAcycA | This study |
| 3612 | CFT073 dsdA cycA | This study |
Mutants were complemented with the relevant gene inserted into low-copy-number plasmid pACYC177 between the BamHI (5′) and BglI (3′) restriction sites. Wild-type E. coli strain CFT073 genomic DNA was amplified using Deep Vent polymerase (New England Biolabs). Creation of the dsdX PCR fragment was carried out using oligos 1171 (5′ GTGGATCCATATGCACTCTCAAATCTGGG 3′) and 1417 (5′ CCAGCCAGCCGGAAGGGCTTAGATGATAAATGACAG 3′). The cycA fragment was generated using oligo 1432 (5′ GCGGTGGATCCATATGGTAGATCAGGTAAAAGTCG 3′) and oligo 1433 (5′ CCAGCCAGCCGGAAGGGCTTATTTCCGCAGTTCAGC 3′). Vectors and inserts were digested with the appropriate enzymes, and vectors were also treated with shrimp alkaline phosphatase (Promega). Ligations were performed with T4 ligase (Promega). Recombinant plasmids were introduced into WAM 2603 by electroporation. Recovered plasmid DNA was screened by NdeI (New England Biolabs) digestion, and the DNA sequences of the inserts were determined to assure the presence of the desired DNA insert. The relevant recombinant plasmids were then introduced into the desired background strain(s).
Genetic screen.
Strains of interest were grown at 37°C overnight on MOPS agar medium with various carbon sources. The agar plates consisted of MOPS medium with one of the following for the carbon source: no carbon source, 43.4 mM glycerol plus 4.76 mM d-serine, 4.76 mM d-serine, or 4.76 mM d-alanine. Cultures were scored for growth or no growth based on comparisons to wild-type CFT073 under each condition. Growth on all varieties of MOPS medium was either equal to that of wild-type CFT073 or absent and scored accordingly as “+” or “−.”
d-Serine deaminase activity assay.
Strains were grown overnight on either MOPS glycerol agar medium or in 2 ml MOPS glycerol broth. Twenty-five-milliliter MOPS broth cultures were inoculated with either a single colony or 250 μl of the overnight broth cultures and incubated at 37°C with shaking until they reached an optical density at 600 nm (OD600) of 0.5. The culture was then split into two 12.5-ml cultures. A 500-μg/ml final concentration of d-serine was added to one culture to induce maximal expression of dsdA, and an equal volume of water was added to the negative control culture. One-milliliter aliquots of each culture were removed at 0, 15, and 30 min post-d-serine addition, the cells were pelleted by centrifugation, the supernatant was discarded, and the cells were flash-frozen in a dry ice and ethanol bath. An additional aliquot of culture was taken to make dilution plates so that activity could be adjusted for CFU present in the sample. The cell pellets were resuspended in 1 ml 1 M KPO4. Fifty microliters of the suspension was added to a glass test tube containing an additional 250 μl of KPO4 and 10 μl of Pop Culture lysis solution (Novagen), with two tubes per cell pellet. Tubes were incubated at 37°C for 15 min to lyse cells. Tubes were then treated with 100 μl of either 9.5 mM d-serine or 100 μl H2O. Tubes were incubated at 37°C for 20 min. Samples were then treated with 0.9 ml of a 3.89 M 2,4-dinitrophenylhydrazine solution (2,4-dinitrophenylhydrazine dissolved in 1.2 N HCl). Samples were incubated at room temperate for 20 min. Reactions were stopped by addition of 1.7 ml 2.5 N NaOH. The OD520 of each sample was measured, and the final optical density of the assay mixture correlates in a linear fashion to pyruvate concentration over an optical density of 0 to 1.5. Because DsdA converts d-serine to ammonia and pyruvate, the presence of pyruvate is a function of DsdA activity within the assay (17).
d-Serine transport.
A 2-ml overnight MOPS medium culture of each strain was inoculated into 25 ml fresh MOPS medium (plus appropriate selective agent as needed) and grown at 37°C with shaking until the culture entered mid-log phase, an OD600 of ∼0.5. Cells were pelleted by centrifugation and washed two times with MOPS-Tris buffer at 4°C. Washed cells were resuspended in 2 ml survival buffer, giving a total protein concentration of approximately 0.3 mg/ml. The cell suspension was allowed to equilibrate to 37°C for 5 min, at which point chloramphenicol was added to a final concentration of 50 μg/ml, and a 50-μl aliquot was removed and stored at −80°C for subsequent bicinchoninic acid assay to measure actual protein concentration. The chloramphenicol-treated cells were incubated for an additional 15 min to ensure cessation of protein synthesis. The cell suspension was split into 50-μl aliquots. Ten microliters of d-[14C]serine (5.5 Ci/mol; 0.6 mM) was added to the suspensions, giving a final concentration of 0.1 mM d-[14C]serine, and allowed to incubate for various times. Five milliliters of MOPS-Tris buffer was added to the cultures to stop transport, and the suspension was filtered through a 0.45-μm-pore-size nitrocellulose membrane filter. The filter was washed with an additional 5 ml of MOPS-Tris buffer. Filters were allowed to dry and then were placed in scintillation vials containing 3 ml Biosafe counting cocktail. The samples were counted in a Beckman scintillation counter. All disintegrations-per-minute values determined by sample counting were normalized to the activity of d-[14C]serine (5.5 Ci/mol) and to the protein concentration of the sample as determined by bicinchoninic acid assay to express counted signal in terms of nmol d-serine · mg total protein−1 · minute−1.
Kinetic analysis was accomplished by incubating cell suspensions with increasing concentrations of d-[14C]serine for 20 s. To examine the dependence of d-serine transport on membrane potential, samples were pretreated with 10 μM carbonyl cyanide m-chlorophenylhydrazone (CCCP) for 3 minutes prior to the start of the transport assay. Data were plotted in Prism 4 (GraphPad Software, San Diego, CA), and apparent Km and Vmax values were determined using the included Michaelis-Menten linear regression template. All uptake data are the result of experiments performed in triplicate unless otherwise indicated.
Cell suspensions for inhibition studies were prepared as described above with the following changes. Prior to addition of d-[14C]serine, the suspensions were pretreated with 10 μl of the desired unlabeled competitor or MOPS-Tris for the control. Amino acids were used at a concentration range of 0.6 to 100 mM to account for the fact that a bacterium in the urinary tract would experience varied concentrations of amino acids (11, 24, 29). Amino acids screened included l-serine, d-alanine, glycine, d-threonine, and d-cycloserine. Additionally, pretreatment with d-cycloserine, CCCP, or a mixture of d-cyloserine and CCCP was used to examine nonspecific binding versus uptake. Preincubation with inhibitor was carried out for 3 min prior to addition of d-[14C]serine. Incubation with d-[14C]serine was allowed to proceed for 3 min and then terminated as with the general uptake experiment. Values obtained from incubation with inhibitors were expressed as a percent inhibition relative to the control that was preincubated with 10 μl MOPS-Tris buffer prior to the addition of d-[14C]serine.
RESULTS
Genetic analysis of DsdX function.
CFT073 deletion mutants of dsdX, dsdA, and cycA and plasmid-complemented strains were created to assess the role of dsdX in d-serine transport and for the ability to grow in the presence of different carbon sources structurally similar to d-serine. Strains were inoculated onto MOPS minimal medium agar plates containing the following as carbon sources: glycerol plus d-serine, d-serine alone, or d-alanine. Inoculated plates were incubated for up to 2.5 days until significant growth of wild-type inoculum (>3-mm colony diameter) was observed. Wild-type CFT073, the positive control, was able to grow under all conditions screened. The growth phenotypes are presented in Table 2.
TABLE 2.
Growth of strains on MOPS minimal medium plates containing different carbon sources
| Strain | Genotype | Growth on plates containinga:
|
||
|---|---|---|---|---|
| d-Serine + glycerol | d-Serine | d-Alanine | ||
| WAM 2267 | Wild-type CFT073 | + | + | + |
| WAM 2614 | dsdA | − | − | + |
| WAM 2719 | cycA | + | + | − |
| WAM 2966 | dsdX | + | + | + |
| WAM 2967 | dsdXA | − | − | + |
| WAM 3147 | dsdX cycA | + | − | − |
| WAM 3160 | dsdXA cycA | + | − | − |
| WAM 3387 | dsdX cycA + pAAdsdX | + | + | − |
| WAM 3388 | dsdXA cycA + pAAdsdX | − | − | − |
| WAM 3481 | dsdX cycA + pAAcycA | + | + | + |
| WAM 3482 | dsdXA cycA + pAAcycA | − | − | + |
+, growth was observed; −, no growth was observed.
The CFT073 dsdX mutant showed the same carbon utilization profile as wild-type CFT073 and was not sensitive to d-serine inhibition. Each strain with a cycA deletion failed to grow on d-alanine as a sole carbon source. The CFT073 dsdX cycA mutant failed to grow on d-serine or d-alanine as a sole carbon source. However, on the d-serine plus glycerol medium, CFT073 dsdX cycA could grow, which suggests that neither d-serine nor d-alanine is able to enter the dsdX cycA mutant cells. The CFT073 dsdXA cycA mutant was able to grow on d-serine plus glycerol. This is a growth condition normally inhibitory to CFT073 dsdA mutant cells, because without dsdA, d-serine is not catabolized and remains toxic to E. coli. Both the CFT073 dsdX and CFT073 cycA strains possess wild-type DsdA enzymatic activity, whereas the CFT073 dsdX cycA mutant lacks DsdA enzymatic activity (data not shown). These data suggest that d-serine is unable to be taken up by CFT073 once both the dsdX and cycA genes are deleted. The addition of plasmid-encoded dsdX to CFT073 dsdX cycA resulted in a resumption of growth on d-serine as a sole carbon source but did not restore growth on d-alanine. Complementation of CFT073 dsdX cycA with cycA resulted in a resumption of growth on d-serine as well as d-alanine. Complementation of CFT073 dsdXA cycA with dsdX or cycA did not result in the ability to grow on either d-serine or d-serine plus glycerol. In order for CFT073 to grow on d-serine as a sole carbon source, dsdX or cycA must be present in addition to dsdA. Loss of cycA leads to an inability to grow on d-alanine as a sole carbon source, but loss of dsdX has no effect on growth in the presence of d-alanine. The growth experiments indicate that DsdX partially overlaps in function with CycA.
d-[14C]serine uptake.
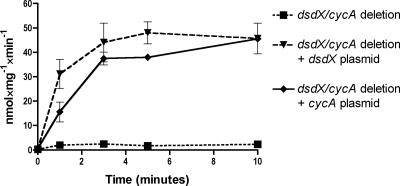
To confirm the conclusions made from the genetic experiments, uptake of d-[14C]serine was monitored. For laboratory E. coli strain K-12, dsdX and cycA genes are expressed under different growth conditions (23, 40). Expression of cycA is induced by low nitrogen levels via the nitrogen-scavenging system (Nac) (40) or σ54 (27); however, dsdX expression is positively controlled by DsdC and the coeffector, d-serine (23). In order to assess DsdX- and CycA-specific d-serine transport under uniform growth conditions, the uptake studies were carried out in the CFT073 dsdX cycA mutant strain complemented with either a plasmid carrying dsdX or cycA under control of the kan promoter. As shown in Fig. 1, both DsdX and CycA were able to transport d-[14C]serine in a time-dependent fashion. The CFT073 dsdX cycA strain fails to transport d-serine, indicating that DsdX and CycA are the only d-serine permeases active under conditions tested. The uptake of d-serine over time in the plasmid-complemented dsdX cycA strains was similar to the uptake of d-serine by the native, chromosomally encoded genes (data not shown); the plasmid-complemented dsdX cycA strains were used for our studies because they express DsdX or CycA under uniform growth conditions. In the DsdX recombinant background, d-serine uptake reached the end point more rapidly than occurs for the CycA-positive strain, although for both strains, the endpoints of d-serine uptake were similar. These data suggest that DsdX was slightly more efficient at d-serine transport than CycA.
FIG. 1.
Time-dependent d-serine uptake. Samples were incubated with 0.1 mM d-[14C]serine (5.5 Ci/mol) for the indicated times. Uptake was stopped with addition of an excess of buffer, and samples were filtered and measured.
Kinetic analysis of d-serine uptake.
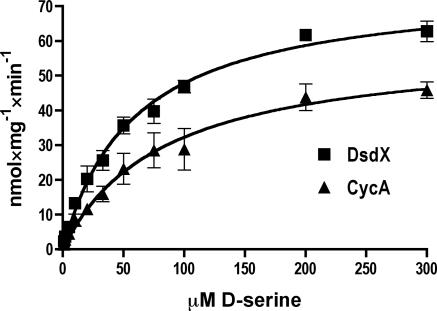
In order to further compare DsdX and CycA function, kinetic experiments of d-serine transport mediated by either DsdX or CycA were conducted. The uptake experiments were similar to the time course evaluations of d-serine transport described above. The principle difference was that the cell suspensions were incubated with d-[14C]serine over a range of concentrations from 1.8 mM to 8 μM at a single time point of 20 s. These results are presented in Fig. 2. The apparent Km for DsdX is 58.75 μM d-serine, and the apparent Vmax is 75.96 nmol · mg−1 · min−1. The apparent Km for CycA is 82.40 μM d-serine, and the apparent Vmax is 58.90 nmol · mg−1 · min−1. Pretreatment of the DsdX-complemented CFT073 dsdX cycA strain with CCCP led to a cessation of d-serine uptake (data not shown).
FIG. 2.
Kinetic evaluation of d-serine transport. Samples were treated with the indicated concentration of d-[14C]serine for 20 s, then the reaction was stopped with an addition of excess buffer, and the sample was filtered and measured. DsdX had an apparent Km of 58.75 μM and an apparent Vmax of 75.96 nmol · mg−1 · min−1. CycA had an apparent Km of 82.40 μM and an apparent Vmax of 58.90 nmol · mg−1 · min−1.
Competitors of d-serine uptake.
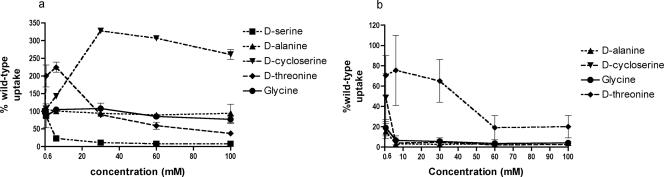
In order to examine other possible substrates of DsdX and CycA uptake, excess potential unlabeled competitors were preincubated for 3 minutes with the CFT073 dsdX cycA mutant strain complemented with either plasmid-carried dsdX or cycA. Cell suspensions of the two strains were then incubated with d-[14C]serine for an additional 3 minutes, and samples were washed and measured as with the uptake experiments described above. The results are expressed as the ratio of labeled d-serine uptake in samples incubated with potential inhibitors to the amount of d-serine taken up without inhibitor. First, pretreatment with unlabeled d-serine was examined. As was expected in both DsdX- and CycA-specific backgrounds, unlabeled d-serine led to a decrease in the uptake of d-[14C]serine in a concentration-dependent fashion. Additional compounds examined for an ability to compete with d-serine for uptake by DsdX included l-serine, d-alanine, d-threonine, glycine, and d-cycloserine. The possible inhibitors of uptake were selected on the basis of known CycA activities (2, 3, 13, 33, 36, 37) or as possible substrates for DsdA (18), the d-serine deaminase. l-Serine and glycine did not interfere with DsdX-mediated transport of d-serine (data not shown). Shown in Fig. 3a are the results of the DsdX-specific competition experiments. d-Threonine, at concentrations greater than 60 mM, caused a slight decrease in DsdX-mediated d-serine transport. d-Alanine did not alter d-serine transport mediated by DsdX. Interestingly, d-cycloserine enhanced DsdX-mediated uptake of labeled d-serine. This is in direct contrast with d-cycloserine inhibition of d-serine uptake by CycA, as shown in Fig. 3b. In the CycA strain, d-[14C]serine uptake was reduced by d-alanine, glycine, and d-cycloserine. Uptake was not inhibited by l-serine or d-threonine. These results are in agreement with previous observations of CycA activity (3, 36) and provide a fair comparison of DsdX function to CycA.
FIG. 3.
Substrate analysis. Samples were preincubated with the indicated amino acid for 3 min, at which time d-[14C]serine was added and the mixture was allowed to incubate for an additional 3 min. Data are plotted as the percentage of wild-type uptake. Percent wild-type uptake of ∼100, no effect; <100, inhibitor; >100, enhancer of transport. (a) The dsdX-complemented strain; (b) the cycA-complemented strain.
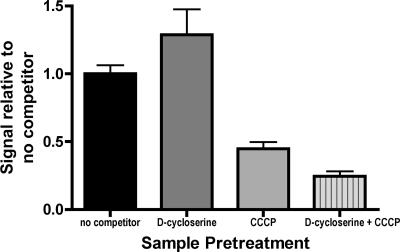
In order to assess if d-cyloserine is transported by DsdX as well as CycA, various plasmid-complemented CFT073 mutants were screened on MOPS minimal agar plates containing glycerol, d-serine, and d-cycloserine. The results of these growth studies are summarized in Table 3. CFT073 expressing CycA alone was required for d-cycloserine growth inhibition. This suggests that the increase in DsdX-mediated d-serine uptake in the presence of d-cycloserine occurred as an interaction independent of d-cycloserine uptake. In order to determine if the d-cycloserine effect on d-serine uptake is related to transport or nonspecific d-serine binding to the cell surface, cell suspensions were treated with d-cycloserine, CCCP, or d-cycloserine and CCCP (Fig. 4). d-Cycloserine alone increases uptake, CCCP reduces uptake, and d-cycloserine plus CCCP further reduces uptake. These data suggest that the effect of d-cycloserine is dependent upon the H+ gradient and is unlikely to be an artifact of enhanced binding of d-serine to the surface of the cell.
TABLE 3.
Growth in the presence of d-cycloserinea
| Strain | Genotype | Growth |
|---|---|---|
| WAM 3387 | dsdX cycA + pAAdsdX | + |
| WAM 3388 | dsdXA cycA + pAAdsdX | − |
| WAM 3481 | dsdX cycA + pAAcycA | − |
| WAM 3482 | dsdXA cycA + pAAcycA | − |
Strains were plated on MOPS with d-serine and d-cycloserine.
FIG. 4.
The d-cycloserine effect requires a H+ gradient. Samples were pretreated as indicated with 60 mM d-cycloserine and/or 10 mM CCCP for 3 minutes. Samples were then exposed to d-[14C]serine for an additional 3 min. The ratio of signal with pretreatment to signal with no pretreatment is plotted.
DISCUSSION
We have shown that DsdX is a d-serine-specific transport protein in E. coli CFT073, sharing some functionality with the CycA permease. Our study of DsdX function began with a deletion of dsdX that did not affect dsdA expression nor disrupt the dsdXA promoter. The loss of dsdX did not cause any detectable change in d-serine catabolism relative to that seen in the wild-type CFT073 strain. It was only when cycA, a multifunctional d-amino acid permease encoding the only previously described d-serine transporter in E. coli, was deleted along with dsdX that the ability to use d-serine was lost.
Our genetic experiments suggest that DsdX is capable of transporting d-serine. Mutation of both dsdX and cycA in CFT073 results in an inability to grow on d-serine or d-alanine as a sole carbon source. Additionally, a CFT073 dsdXA cycA strain is not sensitive to inhibitory concentrations of d-serine despite the lack of an enzyme to degrade d-serine. Loss of cycA alone results in an inability to grow on d-alanine as a sole carbon source, which is in agreement with previous observations that cycA is the sole d-alanine transporter (33).
Both DsdX and CycA are able to transport d-[14C]serine in a time-dependent fashion. DsdX is the more efficient transporter of d-serine, with an apparent Km of 58.75 μM d-serine and an apparent Vmax of 75.96 nmol · mg−1 · min−1. The apparent Km for CycA is 82.40 μM d-serine, and the apparent Vmax is 58.90 nmol · mg−1 · min−1. Whereas CycA is a d-amino acid permease capable of transporting several d-amino acids (2, 3, 36), DsdX is only able to transport d-serine efficiently.
Several observations suggest that DsdX is a H+ symporter. The amino acid sequence of DsdX is similar to that of gluconate transporters, which are known H+ symport permeases (25). Additionally, there is no K+ or Na+ in the MOPS-Tris buffer used in the uptake experiments, only H+, which suggests that DsdX is a H+ symporter. Finally, addition of CCCP, which can disrupt the H+ gradient, leads to a cessation of DsdX-mediated d-serine transport. The CCCP result shows that DsdX requires a H+ gradient to transport d-serine.
The redundancy in E. coli for proteins that transport d-serine is notable. d-Serine is known to be inhibitory, and d-enantiomers of amino acids are not commonly found in bacteria, outside of peptidoglycan. We have previously observed that the loss of the CFT073 d-serine deaminase activity leads to cells that appear to swell when grown in human urine (28). It has also been shown in vitro that d-serine can be recognized by d-alanine-d-alanine ligase, which suggests that d-serine can interfere with bacterial cell wall synthesis (32). A CFT073 dsdA mutant produced pleomorphic cell shapes when grown in human urine in which d-serine concentrations ranged from 5 to 40 μg/ml (11). However, aside from the need to detoxify d-serine, this amino acid may be a primary carbon and nitrogen source in certain environments (28, 29). Extraintestinal E. coli isolates appear more likely to possess a fully functional dsdCXA locus than the diarrheal pathotypes, which often have a truncation in the dsdCXA locus and substitution with genes for sucrose utilization (Moritz and Welch, submitted). Sucrose is readily available in the intestine, particularly for individuals eating diets rich in plant material. Bacteria residing exclusively in the intestine would take advantage of sucrose as a readily catabolizable carbon and energy source. In contrast, bacteria able to grow in the urinary tract are growing in a sugar-poor, amino acid-rich environment. It appears that UPEC evolved to take advantage of a continuous source of significant quantities of d-serine for growth while simultaneously gaining a competitive advantage through detoxification of a growth inhibitor (11).
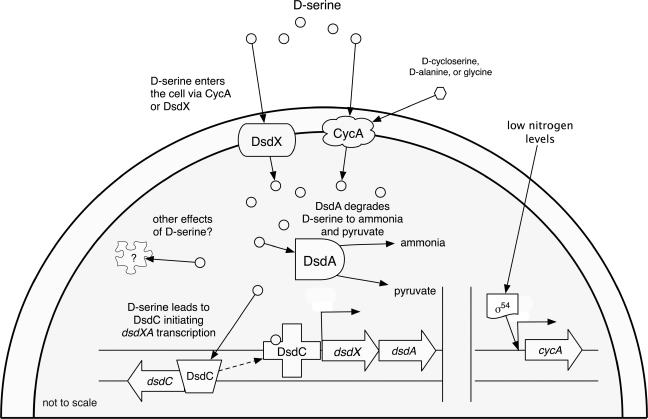
A model pooling past knowledge of d-serine metabolism with our current findings can be found in Fig. 5. This model integrates DsdX and CycA functions on the basis of their differences in expression and the amino acids they transport. CycA is under σ54 control and plays a role as a nitrogen scavenger (33, 40), capable of taking up a range of d-amino acids, including d-alanine and d-serine (2). CycA is the only d-alanine permease that E. coli possesses (33), and so it is important for peptidoglycan synthesis. Uptake of d-serine at low concentrations and at times of nitrogen starvation likely begins with CycA. Once sufficient d-serine enters the cell and interacts with DsdC, dsdXA expression is induced. The subsequent increase in pyruvate and ammonia concentrations are expected to decrease CycA expression, and DsdX then becomes the primary mode of d-serine entry into the cell due to its lower Km and higher Vmax than CycA.
FIG. 5.
Working model of the dsdCXA locus and d-serine metabolism. Both DsdX and CycA are predicted to lie in the inner membrane (5), and our observations are in agreement with this prediction. cycA transcription is controlled by σ54 and Nac (27, 40). Once d-serine enters the cell, it interacts with DsdC, which then induces dsdXA expression. d-Serine is degraded to pyruvate and ammonia by DsdA.
We propose the following model for the regulatory significance of UPEC d-serine catabolism during growth in the urinary tract. Tryptone broth is similar to urine in that it is a carbohydrate-poor but amino acid-rich medium. Compared to other amino acids present in tryptone broth, serine is consumed preferentially by E. coli, which rapidly generates pyruvate and acetate (26). The constant supply of d-serine in urine excreted by the kidneys would lead to relatively constant pyruvate production and continuous acetate secretion by UPEC. The failure to reassimilate acetate as a growth substrate because of micturation would prevent the acetate switch, as described by Wolfe (39). In that metabolic state, it is hypothesized that elevated acetyl phosphate levels are a major regulatory signal for UPEC, leading to positive expression of type 1 fimbriae and capsule (39). This model is currently being tested in our laboratory.
Acknowledgments
We thank Brian Haugen and Holly Hamilton for advice on experiments and manuscript preparation, Shai Pellet for assistance with radiolabeled compounds, Paula Roesch for assistance with mutant construction, and Michael Thomas for discussions on enzyme kinetics.
This research was supported by NIH grant R01DK063250.
REFERENCES
- 1.Blomfield, I. C., M. S. McClain, and B. I. Eisenstein. 1991. Type 1 fimbriae mutants of Escherichia coli K12: characterization of recognized afimbriate strains and construction of new fim deletion mutants. Mol. Microbiol. 5:1439-1445. [DOI] [PubMed] [Google Scholar]
- 2.Cosloy, S. D., and E. McFall. 1973. Metabolism of d-serine in Escherichia coli K-12: mechanism of growth inhibition. J. Bacteriol. 114:685-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cosloy, S. D. 1973. d-Serine transport system in Escherichia coli K-12. J. Bacteriol. 114:679-684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cronan, J. E. 1980. Beta-alanine synthesis in Escherichia coli. J. Bacteriol. 141:1291-1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Daley, D. O., M. Rapp, E. Granseth, K. Melen, D. Drew, and G. von Heijne. 2005. Global topology analysis of the Escherichia coli inner membrane proteome. Science 308:1321-1323. [DOI] [PubMed] [Google Scholar]
- 6.Datsenko, K. A., and B. L. Wanner. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. USA 97:6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Durham, N. N., C. D. Jacobs, and D. Ferguson. 1964. Relationship of a beta-alanine-pyruvic aminotransferase to reversal of d-serine inhibition of growth. J. Bacteriol. 88:1525-1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erikson, O., M. Hertzberg, and T. Näsholm. 2005. The dsdA gene from Escherichia coli provides a novel selectable marker for plant transformation. Plant Mol. Biol. 57:425-433. [DOI] [PubMed] [Google Scholar]
- 9.Hama, H., T. Shimamoto, M. Tsuda, and T. Tsuchiya. 1988. Characterization of a novel l-serine transport system in Escherichia coli. J. Bacteriol. 170:2236-2239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hua, Q., C. Yang, T. Oshima, H. Mori, and K. Shimizu. 2004. Analysis of gene expression in Escherichia coli in response to changes of growth-limiting nutrient in chemostat cultures. Appl. Environ. Microbiol. 70:2354-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang, Y., T. Nishikawa, K. Satoh, T. Iwata, T. Fukushima, T. Santa, H. Homma, and K. Imai. 1998. Urinary excretion of D-serine in human: comparison of different ages and species. Biol. Pharm. Bull. 21:156-162. [DOI] [PubMed] [Google Scholar]
- 12.Jahreis, K., L. Bentler, J. Bockmann, S. Hans, A. Meyer, J. Siepelmeyer, and J. W. Lengeler. 2002. Adaptation of sucrose metabolism in the Escherichia coli wild-type strain EC3132. J. Bacteriol. 184:5307-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kaback, H. R., and A. B. Kostellow. 1968. Glycine uptake in Escherichia coli. I. Glycine uptake by whole cells of Escherichia coli W+ and a D-serine-resistant. J. Biol. Chem. 243:1384-1389. [PubMed] [Google Scholar]
- 14.Krystal, J. H., and D. C. D'Souza. 1998. D-serine and the therapeutic challenge posed by the N-methyl-D-aspartate antagonist model of schizophrenia. Biol. Psychiatry 44:1075-1076. [DOI] [PubMed] [Google Scholar]
- 15.Maas, W. K., and B. D. Davis. 1950. Pantothenate studies. I. Interference by d-serine and l-aspartic acid with pantothenate synthesis in Escherichia coli. J. Bacteriol. 60:733-745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maas, W. K., R. Maas, and E. McFall. 1995. d-Serine deaminase is a stringent selective marker in genetic crosses. J. Bacteriol. 177:459-461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McFall, E. 1975. Escherichia coli K-12 mutant forming a temperature-sensitive d-serine deaminase. J. Bacteriol. 121:1074-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Metzler, D. E., and E. E. Snell. 1952. Deamination of serine. II. d-Serine dehydrase, a vitamin B6 enzyme from Escherichia coli. J. Biol. Chem. 198:363-373. [PubMed] [Google Scholar]
- 19.Mobley, H. L., D. M. Green, A. L. Trifillis, D. E. Johnson, G. R. Chippendale, C. V. Lockatell, B. D. Jones, and J. W. Warren. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 58:1281-1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mothet, J. P., A. T. Parent, H. Wolosker, R. O. J. Brady, D. J. Linden, C. D. Ferris, M. A. Rogawski, and S. H. Snyder. 2000. D-serine is an endogenous ligand for the glycine site of the N-methyl-D-aspartate receptor. Proc. Natl. Acad. Sci. USA 97:4926-4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mothet, J. P. 2001. Physiological relevance of endogenous free D-serine in the mammalian brain: are scientists on a royal road for the treatment of glutamatergic-related brain disorders?. Pathol. Biol. (Paris). 49:655-659. [DOI] [PubMed] [Google Scholar]
- 22.Neidhardt, F. C., P. L. Bloch, and D. F. Smith. 1974. Culture medium for enterobacteria. J. Bacteriol. 119:736-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Norregaard-Madsen, M., E. McFall, and P. Valentin-Hansen. 1995. Organization and transcriptional regulation of the Escherichia coli K-12 d-serine tolerance locus. J. Bacteriol. 177:6456-6461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pätzold, R., A. Schieber, and H. Brückner. 2005. Gas chromatographic quantification of free D-amino acids in higher vertebrates. Biomed. Chromatogr. 19:466-473. [DOI] [PubMed] [Google Scholar]
- 25.Peekhaus, N., S. Tong, J. Reizer, M. H. Saier, E. Murray, and T. Conway. 1997. Characterization of a novel transporter family that includes multiple Escherichia coli gluconate transporters and their homologues. FEMS Microbiol. Lett. 147:233-238. [DOI] [PubMed] [Google Scholar]
- 26.Prüss, B. M., and P. Matsumura. 1996. A regulator of the flagellar regulon of Escherichia coli, flhD, also affects cell division. J. Bacteriol. 178:668-674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reitzer, L., and B. L. Schneider. 2001. Metabolic context and possible physiological themes of σ54-dependent genes in Escherichia coli. Microbiol. Mol. Biol. Rev. 65:422-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Roesch, P. L., P. Redford, S. Batchelet, R. L. Moritz, S. Pellett, B. J. Haugen, F. R. Blattner, and R. A. Welch. 2003. Uropathogenic Escherichia coli use d-serine deaminase to modulate infection of the murine urinary tract. Mol. Microbiol. 49:55-67. [DOI] [PubMed] [Google Scholar]
- 29.Roos, V., M. A. Schembri, G. C. Ulett, and P. Klemm. 2006. Asymptomatic bacteriuria Escherichia coli strain 83972 carries mutations in the foc locus and is unable to express F1C fimbriae. Microbiology 152:1799-1806. [DOI] [PubMed] [Google Scholar]
- 30.Russell, R. R. 1972. Mapping of a d-cycloserine resistance locus in Escherichia coli K-12. J. Bacteriol. 111:622-624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saier, M. H., C. V. Tran, and R. D. Barabote. 2006. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 34:D181-D186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sato, M., K. Kirimura, and K. Kino. 2005. d-Amino acid dipeptide production utilizing d-alanine-d-alanine ligases with novel substrate specificity. J. Biosci. Bioeng. 99:623-628. [DOI] [PubMed] [Google Scholar]
- 33.Schneider, F., R. Krämer, and A. Burkovski. 2004. Identification and characterization of the main beta-alanine uptake system in Escherichia coli. Appl. Microbiol. Biotechnol. 65:576-582. [DOI] [PubMed] [Google Scholar]
- 34.Stevens, E. R., M. Esguerra, P. M. Kim, E. A. Newman, S. H. Snyder, K. R. Zahs, and R. F. Miller. 2003. D-serine and serine racemase are present in the vertebrate retina and contribute to the physiological activation of NMDA receptors. Proc. Natl. Acad. Sci. USA 100:6789-6794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vorachek-Warren, M. K., and J. H. McCusker. 2004. DsdA (D-serine deaminase): a new heterologous MX cassette for gene disruption and selection in Saccharomyces cerevisiae. Yeast 21:163-171. [DOI] [PubMed] [Google Scholar]
- 36.Wargel, R. J., C. A. Hadur, and F. C. Neuhaus. 1971. Mechanism of d-cycloserine action: transport mutants for d-alanine, d-cycloserine, and glycine. J. Bacteriol. 105:1028-1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wargel, R. J., C. A. Shadur, and F. C. Neuhaus. 1970. Mechanism of d-cycloserine action: transport systems for d-alanine, d-cycloserine, l-alanine, and glycine. J. Bacteriol. 103:778-788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Welch, R. A., V. Burland, G. Plunkett, P. Redford, P. Roesch, D. Rasko, E. L. Buckles, S. R. Liou, A. Boutin, J. Hackett, D. Stroud, G. F. Mayhew, D. J. Rose, S. Zhou, D. C. Schwartz, N. T. Perna, H. L. Mobley, M. S. Donnenberg, and F. R. Blattner. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. USA 99:17020-17024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wolfe, A. J. 2005. The acetate switch. Microbiol. Mol. Biol. Rev. 69:12-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zimmer, D. P., E. Soupene, H. L. Lee, V. F. Wendisch, A. B. Khodursky, B. J. Peter, R. A. Bender, and S. Kustu. 2000. Nitrogen regulatory protein C-controlled genes of Escherichia coli: scavenging as a defense against nitrogen limitation. Proc. Natl. Acad. Sci. USA 97:14674-14679. [DOI] [PMC free article] [PubMed] [Google Scholar]