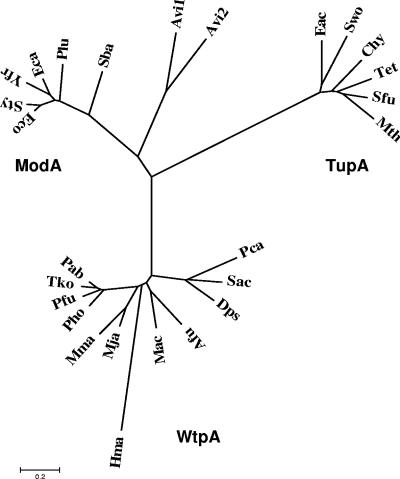
FIG. 4.
Unrooted phylogenetic tree of molybdate and tungstate periplasmic binding proteins. The alignment was made with the 11 homologues (P < 10−20) of the P. furiosus WtpA protein and the five closest homologues, each of the E. coli ModA protein, the E. acidaminophilum TupA protein, and the two A. vinelandii ModA1 and ModA2 proteins. ModA homologues (accession number, identity, similarity): Salmonella enterica serovar Typhimurium LT2 (AE008732.1, 86%, 91%), Erwinia carotovora subsp. atroseptica SCRI1043 (BX950851.1, 69%, 80%), Yersinia frederiksenii ATCC 33641 (NZ_AALE01000012.1, 70%, 80%), Photorhabdus luminescens subsp. laumondii TTO1 (BX571864.1, 61%, 75%), Shewanella baltica OS155 (NZ_AAIO01000008.1, 50%, 71%); TupA homologues (accession number, identity, similarity): Syntrophomonas wolfei strain Goettingen (NZ_AAJG01000003.1, 51%, 67%), Thermoanaerobacter ethanolicus ATCC 33223 (NZ_AAKQ01000001.1, 50%, 70%), Carboxydothermus hydrogenoformans Z-2901 (CP000141.1, 50%, 65%), Syntrophobacter fumaroxidans MPOB (NZ_AAJF01000122.1, 48%, 66%), Moorella thermoacetica ATCC 39073 (NC_007644.1, 46%, 64%); WtpA homologues (accession number, identity, similarity): Thermococcus kodakarensis KOD1 (AP006878.1, 75%, 87%), Pyrococcus abyssi GE5 (AJ248283.1, 74%, 87%), Pyrococcus horikoshii shinkaj OT3 (BA000001.2, 73%, 90%), Methanococcus jannaschii DSM2661 (L77117.1, 49%, 69%), Archaeoglobus fulgidus DSM4304 (NC_000917.1, 46%, 67%), Methanococcus maripaludis S2 (BX957223.1, 46%, 63%), Methanosarcina acetivorans C2A (AE010299.1, 43%, 63%), Syntrophus aciditrophicus SB (NC_007759.1, 39%, 60%), Desulfotalea psychrophila LSv54 (CR522870.1, 40%, 56%), Pelobacter carbinolicus DSM 2380 (CP000142.1, 36%, 55%), Haloarcula marismortui ATCC 43049 (AY596297.1, 29%, 47%).