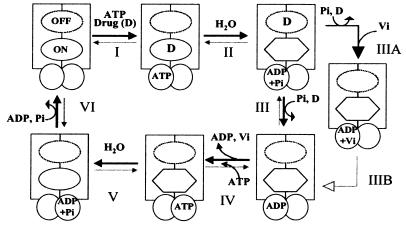
Figure 6.
A proposed scheme for the catalytic cycle of Pgp. The rectangles represent the N and C halves of Pgp and the overlapping circles the two NBS. The ovals represent the substrate binding sites, the on site is depicted by the continuous perimeter, and the off site with a dotted line (see Discussion for an explanation of the on and off sites). The hexagon portrays the on site with reduced affinity for the drug. Step I: Substrate binds to the on site of Pgp, and ATP binds to one or both of the two NBS. Step II: ATP is hydrolyzed and the drug is possibly moved to the lower affinity off site. Step III: Pi is released and the drug extruded from Pgp at this step. Step IIIA: When Pi is replaced by Vi, the Pgp⋅ADP⋅Vi complex is generated, which also exhibits a reduced affinity for substrate. Step IV/step IIIB: The ADP is disassociated; however, the affinity for substrate continues to be low. Step V: After disassociation of the ADP, an additional molecule(s) of ATP is hydrolyzed and the conformation of Pgp is restored to its original state with high affinity for substrate binding. Step VI: The release of Pi, disassociation of ADP followed by the initiation of the next catalytic cycle. It is not known, however, whether release of ADP necessarily precedes the binding of the next molecule of substrate after step VI. ADP disassociation (from the previous cycle), drug binding, and ATP binding presumably could occur in any sequence in steps VI to I. Though, we depict ATP binding and hydrolysis as occurring in the N half of Pgp our data do not suggest any preference in the presence of saturating concentrations of ATP or 8AzidoATP for either of the two sites, and the process is most likely random. However consistent with the alternate site model (4), ATP hydrolysis would occur at a site different from that in step II and therefore is depicted at the NBS in the C half at step V.